Identification of four gene variants associated with myocardial infarction
- PMID: 16175505
- PMCID: PMC1275608
- DOI: 10.1086/491674
Identification of four gene variants associated with myocardial infarction
Abstract
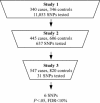
Family history is a major risk factor for myocardial infarction (MI). However, known gene variants associated with MI cannot fully explain the genetic component of MI risk. We hypothesized that a gene-centric association study that was not limited to candidate genes could identify novel genetic associations with MI. We studied 11,053 single-nucleotide polymorphisms (SNPs) in 6,891 genes, focusing on SNPs that could influence gene function to increase the likelihood of identifying disease-causing gene variants. To minimize false-positive associations generated by multiple testing, two studies were used to identify a limited number of nominally associated SNPs; a third study tested the hypotheses that these SNPs are associated with MI. In the initial study (of 340 cases and 346 controls), 637 SNPs were associated with MI (P<.05); these were evaluated in a second study (of 445 cases and 606 controls), and 31 of the 637 SNPs were associated with MI (P<.05) and had the same risk allele as in the first study. For each of these 31 SNPs, we tested the hypothesis that it is associated with MI, using a third study (of 560 cases and 891 controls). We found that four of these gene variants were associated with MI (P<.05; false-discovery rate <10%) and had the same risk allele as in the first two studies. These gene variants encode the cytoskeletal protein palladin (KIAA0992 [odds ratio (OR) 1.40]), a tyrosine kinase (ROS1 [OR 1.75]), and two G protein-coupled receptors (TAS2R50 [OR 1.58] and OR13G1 [OR 1.40]); all ORs are for carriers of two versus zero risk alleles. These findings could lead to a better understanding of MI pathophysiology and improved patient risk assessment.
Figures
References
Web Resources
-
- Applied Biosystems, http://myscience.appliedbiosystems.com/ (for the Celera database)
-
- Authors' Web site, http://www.celeradiagnostics.com/pdf/MI4_variants_supplemental.pdf
-
- International HapMap Project, http://www.hapmap.org/
-
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for KIAA0992 and ROS1)
References
-
- American Heart Association (2002) Heart disease and stroke statistics: 2005 update. American Heart Association, Dallas
-
- Benjamini Y, Hochberg Y (1995) Controlling false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57:1289–1300
-
- Botstein D, Risch N (2003) Discovering genotypes underlying human phenotypes: past successes for Mendelian disease, future approaches for complex disease. Nat Genet Suppl 33:228–237 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

