EXPANDER--an integrative program suite for microarray data analysis
- PMID: 16176576
- PMCID: PMC1261157
- DOI: 10.1186/1471-2105-6-232
EXPANDER--an integrative program suite for microarray data analysis
Abstract
Background: Gene expression microarrays are a prominent experimental tool in functional genomics which has opened the opportunity for gaining global, systems-level understanding of transcriptional networks. Experiments that apply this technology typically generate overwhelming volumes of data, unprecedented in biological research. Therefore the task of mining meaningful biological knowledge out of the raw data is a major challenge in bioinformatics. Of special need are integrative packages that provide biologist users with advanced but yet easy to use, set of algorithms, together covering the whole range of steps in microarray data analysis.
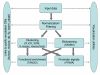
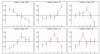
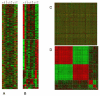
Results: Here we present the EXPANDER 2.0 (EXPression ANalyzer and DisplayER) software package. EXPANDER 2.0 is an integrative package for the analysis of gene expression data, designed as a 'one-stop shop' tool that implements various data analysis algorithms ranging from the initial steps of normalization and filtering, through clustering and biclustering, to high-level functional enrichment analysis that points to biological processes that are active in the examined conditions, and to promoter cis-regulatory elements analysis that elucidates transcription factors that control the observed transcriptional response. EXPANDER is available with pre-compiled functional Gene Ontology (GO) and promoter sequence-derived data files for yeast, worm, fly, rat, mouse and human, supporting high-level analysis applied to data obtained from these six organisms.
Conclusion: EXPANDER integrated capabilities and its built-in support of multiple organisms make it a very powerful tool for analysis of microarray data. The package is freely available for academic users at http://www.cs.tau.ac.il/~rshamir/expander.
Figures



References
-
- Tamayo P, Slonim D, Mesirov J, Zhu Q, Kitareewan S, Dmitrovsky E, Lander ES, Golub TR. Interpreting patterns of gene expression with self-organizing maps: methods and application to hematopoietic differentiation. Proc Natl Acad Sci U S A. 1999;96:2907–2912. doi: 10.1073/pnas.96.6.2907. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

