Comparative genomic analysis of Chlamydia trachomatis oculotropic and genitotropic strains
- PMID: 16177312
- PMCID: PMC1230933
- DOI: 10.1128/IAI.73.10.6407-6418.2005
Comparative genomic analysis of Chlamydia trachomatis oculotropic and genitotropic strains
Abstract
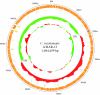
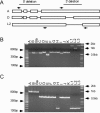
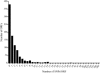
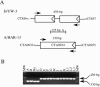
Chlamydia trachomatis infection is an important cause of preventable blindness and sexually transmitted disease (STD) in humans. C. trachomatis exists as multiple serovariants that exhibit distinct organotropism for the eye or urogenital tract. We previously reported tissue-tropic correlations with the presence or absence of a functional tryptophan synthase and a putative GTPase-inactivating domain of the chlamydial toxin gene. This suggested that these genes may be the primary factors responsible for chlamydial disease organotropism. To test this hypothesis, the genome of an oculotropic trachoma isolate (A/HAR-13) was sequenced and compared to the genome of a genitotropic (D/UW-3) isolate. Remarkably, the genomes share 99.6% identity, supporting the conclusion that a functional tryptophan synthase enzyme and toxin might be the principal virulence factors underlying disease organotropism. Tarp (translocated actin-recruiting phosphoprotein) was identified to have variable numbers of repeat units within the N and C portions of the protein. A correlation exists between lymphogranuloma venereum serovars and the number of N-terminal repeats. Single-nucleotide polymorphism (SNP) analysis between the two genomes highlighted the minimal genetic variation. A disproportionate number of SNPs were observed within some members of the polymorphic membrane protein (pmp) autotransporter gene family that corresponded to predicted T-cell epitopes that bind HLA class I and II alleles. These results implicate Pmps as novel immune targets, which could advance future chlamydial vaccine strategies. Lastly, a novel target for PCR diagnostics was discovered that can discriminate between ocular and genital strains. This discovery will enhance epidemiological investigations in nations where both trachoma and chlamydial STD are endemic.
Figures


References
-
- Brunham, R. C., and J. Rey-Ladino. 2005. Immunology of chlamydia infection: implications for a Chlamydia trachomatis vaccine. Nature 5:149-161. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

