Transcriptional repressor DREAM regulates T-lymphocyte proliferation and cytokine gene expression
- PMID: 16177826
- PMCID: PMC1276700
- DOI: 10.1038/sj.emboj.7600810
Transcriptional repressor DREAM regulates T-lymphocyte proliferation and cytokine gene expression
Abstract
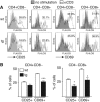
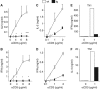
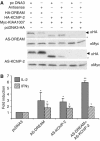
Downstream Regulatory Element Antagonist Modulator (DREAM) is a Ca2+-dependent transcriptional repressor expressed in the brain, thyroid gland and thymus. Here, we analyzed the function of DREAM and the related protein KChIP-2 in the immune system using transgenic (tg) mice expressing a cross-dominant active mutant (EFmDREAM) for DREAM and KChIPs Ca2+-dependent transcriptional derepression. EFmDREAM tg mice showed reduced T-cell proliferation. Tg T cells exhibited decreased interleukin (IL)-2, -4 and interferon (IFN)gamma production after polyclonal activation and following antigen-specific response. Chromatin immunoprecipitation and transfection assays showed that DREAM binds to and represses transcription from these cytokine promoters. Importantly, specific transient knockdown of DREAM or KChIP-2 induced basal expression of IL-2 and IFNgamma in wild-type splenocytes. These data propose DREAM and KChIP-2 as Ca2+-dependent repressors of the immune response.
Figures





References
-
- An WF, Bowlby MR, Betty M, Cao J, Ling HP, Mendoza G, Hinson JW, Mattsson KI, Strassle BW, Trimmer JS, Rhodes KJ (2000) Modulation of A-type potassium channels by a family of Ca2+ sensors. Nature 403: 553–556 - PubMed
-
- Argyropoulos C, Nikiforidis GC, Theodoropoulou M, Adamopoulos P, Boubali S, Georgakopoulos TN, Paliogianni F, Papavassiliou AG, Mouzaki A (2004) Mining microarray data to identify transcription factors expressed in naive resting but not activated T lymphocytes. Genes Immun 5: 16–25 - PubMed
-
- Benedito AB, Lehtinen M, Massol R, Lopes UG, Kirchhausen T, Rao A, Bonni A (2005) The transcription factor NFAT3 mediates neuronal survival. J Biol Chem 280: 2818–2825 - PubMed
-
- Buckley AF, Kuo CT, Leiden JM (2001) Transcription factor LKLF is sufficient to program T cell quiescence via a c-Myc-dependent pathway. Nat Immunol 2: 698–704 - PubMed
-
- Buxbaum JD, Choi EK, Luo Y, Lilliehook C, Crowley AC, Merriam DE, Wasco W (1998) Calsenilin: a Ca2+-binding protein that interacts with the presenilins and regulates the levels of a presenilin fragment. Nat Med 4: 1177–1181 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

