An aneuploid mouse strain carrying human chromosome 21 with Down syndrome phenotypes
- PMID: 16179473
- PMCID: PMC1378183
- DOI: 10.1126/science.1114535
An aneuploid mouse strain carrying human chromosome 21 with Down syndrome phenotypes
Abstract
Aneuploidies are common chromosomal defects that result in growth and developmental deficits and high levels of lethality in humans. To gain insight into the biology of aneuploidies, we manipulated mouse embryonic stem cells and generated a trans-species aneuploid mouse line that stably transmits a freely segregating, almost complete human chromosome 21 (Hsa21). This "transchromosomic" mouse line, Tc1, is a model of trisomy 21, which manifests as Down syndrome (DS) in humans, and has phenotypic alterations in behavior, synaptic plasticity, cerebellar neuronal number, heart development, and mandible size that relate to human DS. Transchromosomic mouse lines such as Tc1 may represent useful genetic tools for dissecting other human aneuploidies.
Figures
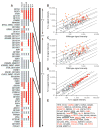

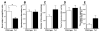
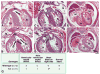
Comment in
-
Genetics. Mouse with human chromosome should boost Down syndrome research.Science. 2005 Sep 23;309(5743):1975. doi: 10.1126/science.309.5743.1975a. Science. 2005. PMID: 16179439 No abstract available.
References
-
- Chrast R, et al. Hum Mol Genet. 2000;9:1853. - PubMed
-
- Smith DJ, et al. Nat Genet. 1997;16:28. - PubMed
-
- Antonarakis SE, Lyle R, Dermitzakis ET, Reymond A, Deutsch S. Nat Rev Genet. 2004;5:725. - PubMed
-
- Nelson DL, Gibbs RA. Science. 2004;306:619. - PubMed
-
- Gardiner K, Fortna A, Bechtel L, Davisson MT. Gene. 2003;318:137. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

