Human-zebrafish non-coding conserved elements act in vivo to regulate transcription
- PMID: 16179648
- PMCID: PMC1236720
- DOI: 10.1093/nar/gki853
Human-zebrafish non-coding conserved elements act in vivo to regulate transcription
Abstract
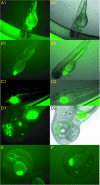
Whole genome comparisons of distantly related species effectively predict biologically important sequences--core genes and cis-acting regulatory elements (REs)--but require experimentation to verify biological activity. To examine the efficacy of comparative genomics in identification of active REs from anonymous, non-coding (NC) sequences, we generated a novel alignment of the human and draft zebrafish genomes, and contrasted this set to existing human and fugu datasets. We tested the transcriptional regulatory potential of candidate sequences using two in vivo assays. Strict selection of non-genic elements which are deeply conserved in vertebrate evolution identifies 1744 core vertebrate REs in human and two fish genomes. We tested 16 elements in vivo for cis-acting gene regulatory properties using zebrafish transient transgenesis and found that 10 (63%) strongly modulate tissue-specific expression of a green fluorescent protein reporter vector. We also report a novel quantitative enhancer assay with potential for increased throughput based on normalized luciferase activity in vivo. This complementary system identified 11 (69%; including 9 of 10 GFP-confirmed elements) with cis-acting function. Together, these data support the utility of comparative genomics of distantly related vertebrate species to identify REs and provide a scaleable, in vivo quantitative assay to define functional activity of candidate REs.
Figures



References
-
- Waterston R.H., Lindblad-Toh K., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. - PubMed
-
- Ovcharenko I., Stubbs L., Loots G.G. Interpreting mammalian evolution using Fugu genome comparisons. Genomics. 2004;84:890–895. - PubMed
-
- Loots G.G., Locksley R.M., Blankespoor C.M., Wang Z.E., Miller W., Rubin E.M., Frazer K.A. Identification of a coordinate regulator of interleukins 4, 13, and 5 by cross-species sequence comparisons. Science. 2000;288:136–140. - PubMed
-
- Pennacchio L.A., Olivier M., Hubacek J.A., Cohen J.C., Cox D.R., Fruchart J.C., Krauss R.M., Rubin E.M. An apolipoprotein influencing triglycerides in humans and mice revealed by comparative sequencing. Science. 2001;294:169–173. - PubMed

