The Sireviruses, a plant-specific lineage of the Ty1/copia retrotransposons, interact with a family of proteins related to dynein light chain 8
- PMID: 16183843
- PMCID: PMC1256001
- DOI: 10.1104/pp.105.065680
The Sireviruses, a plant-specific lineage of the Ty1/copia retrotransposons, interact with a family of proteins related to dynein light chain 8
Abstract
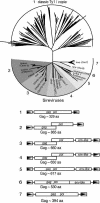
Plant genomes are rich in long terminal repeat retrotransposons, and here we describe a plant-specific lineage of Ty1/copia elements called the Sireviruses. The Sireviruses vary greatly in their genomic organization, and many have acquired additional coding information in the form of an envelope-like open reading frame and an extended gag gene. Two-hybrid screens were conducted with the novel domain of Gag (the Gag extension) encoded by a representative Sirevirus from maize (Zea mays) called Hopie. The Hopie Gag extension interacts with a protein related to dynein light chain 8 (LC8). LC8 also interacts with the Gag extension from a Hopie homolog from rice (Oryza sativa). Amino acid motifs were identified in both Hopie Gag and LC8 that are responsible for the interaction. Two amino acids critical for Gag recognition map within the predicted LC8-binding cleft. Two-hybrid screens were also conducted with the Gag extension encoded by the soybean (Glycine max) SIRE1 element, and an interaction was found with light chain 6 (LC6), a member of the LC8 protein family. LC8 and LC6 proteins are components of the dynein microtubule motor, with LC8 being a versatile adapter that can bind many unrelated cellular proteins and viruses. Plant LC8 and LC6 genes are abundant and divergent, yet flowering plants do not encode other components of the dynein motor. Although, to our knowledge, no cellular roles for plant LC8 family members have been proposed, we hypothesize that binding of LC8 proteins to Gag aids in the movement of retrotransposon virus-like particles within the plant cell or possibly induces important conformational changes in the Gag protein.
Figures






References
-
- Bartel PL, Fields S (1995) Analyzing protein-protein interactions using two-hybrid system. Methods Enzymol 254: 241–263 - PubMed
-
- Boeke JD, Eickbush TH, Sandmeyer SB, Voytas DF (2004) Pseudoviridae. In CM Fauquet, ed, Virus Taxonomy: Eighth Report of the International Committee on Taxonomy of Viruses. Academic Press, New York, pp 663–672
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

