Early effects of salinity on water transport in Arabidopsis roots. Molecular and cellular features of aquaporin expression
- PMID: 16183846
- PMCID: PMC1255996
- DOI: 10.1104/pp.105.065029
Early effects of salinity on water transport in Arabidopsis roots. Molecular and cellular features of aquaporin expression
Abstract
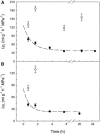
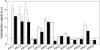
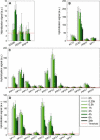
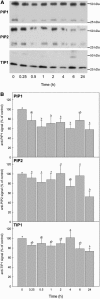
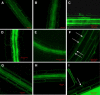
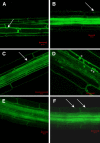
Aquaporins facilitate the uptake of soil water and mediate the regulation of root hydraulic conductivity (Lp(r)) in response to a large variety of environmental stresses. Here, we use Arabidopsis (Arabidopsis thaliana) plants to dissect the effects of salt on both Lp(r) and aquaporin expression and investigate possible molecular and cellular mechanisms of aquaporin regulation in plant roots under stress. Treatment of plants by 100 mm NaCl was perceived as an osmotic stimulus and induced a rapid (half-time, 45 min) and significant (70%) decrease in Lp(r), which was maintained for at least 24 h. Macroarray experiments with gene-specific tags were performed to investigate the expression of all 35 genes of the Arabidopsis aquaporin family. Transcripts from 20 individual aquaporin genes, most of which encoded members of the plasma membrane intrinsic protein (PIP) and tonoplast intrinsic protein (TIP) subfamilies, were detected in nontreated roots. All PIP and TIP aquaporin transcripts with a strong expression signal showed a 60% to 75% decrease in their abundance between 2 and 4 h following exposure to salt. The use of antipeptide antibodies that cross-reacted with isoforms of specific aquaporin subclasses revealed that the abundance of PIP1s decreased by 40% as early as 30 min after salt exposure, whereas PIP2 and TIP1 homologs showed a 20% to 40% decrease in abundance after 6 h of treatment. Expression in transgenic plants of aquaporins fused to the green fluorescent protein revealed that the subcellular localization of TIP2;1 and PIP1 and PIP2 homologs was unchanged after 45 min of exposure to salt, whereas a TIP1;1-green fluorescent protein fusion was relocalized into intracellular spherical structures tentatively identified as intravacuolar invaginations. The appearance of intracellular structures containing PIP1 and PIP2 homologs was occasionally observed after 2 h of salt treatment. In conclusion, this work shows that exposure of roots to salt induces changes in aquaporin expression at multiple levels. These changes include a coordinated transcriptional down-regulation and subcellular relocalization of both PIPs and TIPs. These mechanisms may act in concert to regulate root water transport, mostly in the long term (> or =6 h).
Figures
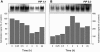
References
-
- Asamizu E, Nakamura Y, Sato S, Tabata S (2000) A large scale analysis of cDNA in Arabidopsis thaliana: generation of 12,028 non-redundant expressed sequence tags from normalized and size-selected cDNA libraries. DNA Res 7: 175–180 - PubMed
-
- Barkla BJ, Vera-Estrella R, Pantoja O, Kirch HH, Bohnert HJ (1999) Aquaporin localization—how valid are the TIP and PIP labels? Trends Plant Sci 4: 86–88 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

