Influence of substrate binding on the mechanical stability of mouse dihydrofolate reductase
- PMID: 16183885
- PMCID: PMC1366864
- DOI: 10.1529/biophysj.105.072066
Influence of substrate binding on the mechanical stability of mouse dihydrofolate reductase
Abstract
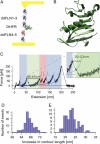
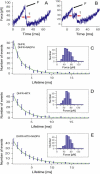
We investigated the effect of substrate binding on the mechanical stability of mouse dihydrofolate reductase using single-molecule force spectroscopy by atomic force microscopy. We find that under mechanical forces dihydrofolate reductase unfolds via a metastable intermediate with lifetimes on the millisecond timescale. Based on the measured length increase of approximately 22 nm we suggest a structure for this intermediate with intact substrate binding sites. In the presence of the substrate analog methotrexate and the cofactor NADPH lifetimes of this intermediate are increased by up to a factor of two. Comparing mechanical and thermodynamic stabilization effects of substrate binding suggests mechanical stability is dominated by local interactions within the protein structure. These experiments demonstrate that protein mechanics can be used to probe the substrate binding status of an enzyme.
Figures
Comment in
-
Fingerprinting DHFR in single-molecule AFM studies.Biophys J. 2006 Sep 1;91(5):2009-10, discussion 2011-2. doi: 10.1529/biophysj.106.085126. Epub 2006 Jun 16. Biophys J. 2006. PMID: 16782796 Free PMC article. No abstract available.
References
-
- Rief, M., M. Gautel, F. Oesterhelt, J. M. Fernandez, and H. E. Gaub. 1997. Reversible unfolding of individual titin immunoglobulin domains by AFM. Science. 276:1109–1112. - PubMed
-
- Marszalek, P. E., H. Lu, H. Li, M. Carrion-Vazquez, A. F. Oberhauser, K. Schulten, and J. M. Fernandez. 1999. Mechanical unfolding intermediates in titin modules. Nature. 402:100–103. - PubMed
-
- Touchette, N. A., K. M. Perry, and C. R. Matthews. 1986. Folding of dihydrofolate reductase from Escherichia coli. Biochemistry. 25:5445–5452. - PubMed
-
- Chunduru, S. K., V. Cody, J. R. Luft, W. Pangborn, J. R. Appleman, and R. L. Blakley. 1994. Methotrexate-resistant variants of human dihydrofolate reductase. Effects of Phe31 substitutions. J. Biol. Chem. 269:9547–9555. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

