Cadherin mechanics and complexation: the importance of calcium binding
- PMID: 16183887
- PMCID: PMC1366956
- DOI: 10.1529/biophysj.105.067322
Cadherin mechanics and complexation: the importance of calcium binding
Abstract
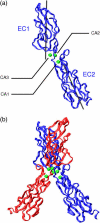
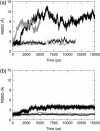
E-cadherins belong to a family of membrane-bound, cellular adhesion proteins. Their adhesive properties mainly involve the two N-terminal extracellular domains (EC1 and EC2). The junctions between these domains are characterized by calcium ion binding sites, and calcium ions are essential for the correct functioning of E-cadherins. Calcium is believed to rigidify the extracellular portion of the protein, which, when complexed, adopts a rod-like conformation. Here, we use molecular dynamics simulations to investigate the dynamics of the EC1-2 portion of E-cadherin in the presence and in the absence of calcium ions. These simulations confirm that apo-cadherin shows much higher conformational flexibility on a nanosecond timescale than the calcium-bound form. It is also shown that although the apo-cadherin fragment can spontaneously complex potassium, these monovalent ions are incapable of rigidifying the interdomain junctions. In contrast, removal of the most solvent-exposed calcium ion at the EC1-2 junction does not significantly perturb the dynamical behavior of the fragment. We have also extended this study to the cis-dimer formed from two EC1-2 fragments, potentially involved in cellular adhesion. Here again, it is shown that the presence of calcium is an important factor in both rigidifying and stabilizing the complex.
Figures







Similar articles
-
Dynamics and stability of E-cadherin dimers.Biophys J. 2006 Dec 1;91(11):3964-71. doi: 10.1529/biophysj.106.087213. Epub 2006 Sep 15. Biophys J. 2006. PMID: 16980367 Free PMC article.
-
Sequential binding of calcium leads to dimerization in neural cadherin.Biochemistry. 2011 Apr 12;50(14):2973-82. doi: 10.1021/bi101872b. Epub 2011 Mar 21. Biochemistry. 2011. PMID: 21366346
-
The allosteric role of the Ca2+ switch in adhesion and elasticity of C-cadherin.Biophys J. 2008 Jun;94(12):4621-33. doi: 10.1529/biophysj.107.125591. Epub 2008 Mar 7. Biophys J. 2008. PMID: 18326636 Free PMC article.
-
Mechanism and dynamics of cadherin adhesion.Annu Rev Biomed Eng. 2006;8:259-87. doi: 10.1146/annurev.bioeng.8.061505.095753. Annu Rev Biomed Eng. 2006. PMID: 16834557 Review.
-
Thinking outside the cell: how cadherins drive adhesion.Trends Cell Biol. 2012 Jun;22(6):299-310. doi: 10.1016/j.tcb.2012.03.004. Epub 2012 May 1. Trends Cell Biol. 2012. PMID: 22555008 Free PMC article. Review.
Cited by
-
Innovative method for quantification of cell-cell adhesion in 96-well plates.Cell Adh Migr. 2011 May-Jun;5(3):215-9. doi: 10.4161/cam.5.3.14648. Epub 2011 May 1. Cell Adh Migr. 2011. PMID: 21339704 Free PMC article.
-
Ganetespib with Methotrexate Acts Synergistically to Impede NF-κB/p65 Signaling in Human Lung Cancer A549 Cells.Pharmaceuticals (Basel). 2023 Feb 2;16(2):230. doi: 10.3390/ph16020230. Pharmaceuticals (Basel). 2023. PMID: 37259378 Free PMC article.
-
Biallelic variants in PCDHGC4 cause a novel neurodevelopmental syndrome with progressive microcephaly, seizures, and joint anomalies.Genet Med. 2021 Nov;23(11):2138-2149. doi: 10.1038/s41436-021-01260-4. Epub 2021 Jul 9. Genet Med. 2021. PMID: 34244665 Free PMC article.
-
Using Coarse-Grained Simulations to Characterize the Mechanisms of Protein-Protein Association.Biomolecules. 2020 Jul 15;10(7):1056. doi: 10.3390/biom10071056. Biomolecules. 2020. PMID: 32679892 Free PMC article.
-
Beyond Cell-Cell Adhesion: Sensational Cadherins for Hearing and Balance.Cold Spring Harb Perspect Biol. 2018 Sep 4;10(9):a029280. doi: 10.1101/cshperspect.a029280. Cold Spring Harb Perspect Biol. 2018. PMID: 28847902 Free PMC article. Review.
References
-
- Yap, A. S., W. M. Brieher, and B. M. Gumbiner. 1997. Molecular and functional analysis of cadherin-based adherens junctions. Annu. Rev. Cell Dev. Biol. 13:119–146. - PubMed
-
- Takeichi, M. 1990. Cadherins: a molecular family important in selective cell-cell adhesion. Annu. Rev. Biochem. 59:237–252. - PubMed
-
- Patel, S. D., C. P. Chen, F. Bahna, B. Honig, and L. Shapiro. 2003. Cadherin-mediated cell-cell adhesion: sticking together as a family. Curr. Opin. Struct. Biol. 13:690–698. - PubMed
-
- Handschuh, G., B. Luber, P. Hutzler, H. Hofler, and K. F. Becker. 2001. Single amino acid substitutions in conserved extracellular domains of E-cadherin differ in their functional consequences. J. Mol. Biol. 314:445–454. - PubMed
-
- Alattia, J. R., J. B. Ames, T. Porumb, K. I. Tong, Y. M. Heng, P. Ottensmeyer, C. M. Kay, and M. Ikura. 1997. Lateral self-assembly of E-cadherin directed by cooperative calcium binding. FEBS Lett. 417:405–408. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

