Extensive chromosomal repatterning and the evolution of sterility barriers in hybrid sunflower species
- PMID: 16183908
- PMCID: PMC1456521
- DOI: 10.1534/genetics.105.042242
Extensive chromosomal repatterning and the evolution of sterility barriers in hybrid sunflower species
Abstract
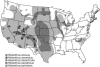
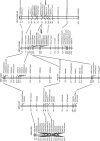
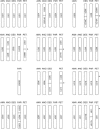
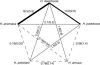
New species may arise via hybridization and without a change in ploidy. This process, termed homoploid hybrid speciation, is theoretically difficult because it requires the development of reproductive barriers in sympatry or parapatry. Theory suggests that isolation may arise through rapid karyotypic evolution and/or ecological divergence of hybrid neospecies. Here, we investigate the role of karyotypic change in homoploid hybrid speciation by generating detailed genetic linkage maps for three hybrid sunflower species, Helianthus anomalus, H. deserticola, and H. paradoxus, and comparing these maps to those previously generated for the parental species, H. annuus and H. petiolaris. We also conduct a quantitative trait locus (QTL) analysis of pollen fertility in a BC2 population between the parental species and assess levels of pollen and seed fertility in all cross-combinations of the hybrid and parental species. The three hybrid species are massively divergent from their parental species in karyotype; gene order differences were observed for between 9 and 11 linkage groups (of 17 total), depending on the comparison. About one-third of the karyoypic differences arose through the sorting of chromosomal rearrangements that differentiate the parental species, but the remainder appear to have arisen de novo (six breakages/six fusions in H. anomalus, four breakages/three fusions in H. deserticola, and five breakages/five fusions in H. paradoxus). QTL analyses indicate that the karyotypic differences contribute to reproductive isolation. Nine of 11 pollen viability QTL occur on rearranged chromosomes and all but one map close to a rearrangement breakpoint. Finally, pollen and seed fertility estimates for F1's between the hybrid and parental species fall below 11%, which is sufficient for evolutionary independence of the hybrid neospecies.
Figures

Similar articles
-
Natural selection for salt tolerance quantitative trait loci (QTLs) in wild sunflower hybrids: implications for the origin of Helianthus paradoxus, a diploid hybrid species.Mol Ecol. 2003 May;12(5):1225-35. doi: 10.1046/j.1365-294x.2003.01803.x. Mol Ecol. 2003. PMID: 12694286
-
Rapid hybrid speciation in wild sunflowers.Proc Natl Acad Sci U S A. 1998 Sep 29;95(20):11757-62. doi: 10.1073/pnas.95.20.11757. Proc Natl Acad Sci U S A. 1998. PMID: 9751738 Free PMC article.
-
Crossing relationships among ancient and experimental sunflower hybrid lineages.Evolution. 2000 Jun;54(3):859-65. doi: 10.1111/j.0014-3820.2000.tb00086.x. Evolution. 2000. PMID: 10937259
-
The ecological genetics of homoploid hybrid speciation.J Hered. 2005 May-Jun;96(3):241-52. doi: 10.1093/jhered/esi026. Epub 2004 Dec 23. J Hered. 2005. PMID: 15618301 Free PMC article. Review.
-
The role of homoploid hybridization in evolution: a century of studies synthesizing genetics and ecology.Am J Bot. 2014 Aug;101(8):1247-58. doi: 10.3732/ajb.1400201. Epub 2014 Aug 12. Am J Bot. 2014. PMID: 25156978 Review.
Cited by
-
Recombining Your Way Out of Trouble: The Genetic Architecture of Hybrid Fitness under Environmental Stress.Mol Biol Evol. 2020 Jan 1;37(1):167-182. doi: 10.1093/molbev/msz211. Mol Biol Evol. 2020. PMID: 31518427 Free PMC article.
-
The Role of Interspecific Hybridisation in Adaptation and Speciation: Insights From Studies in Senecio.Front Plant Sci. 2022 Jun 23;13:907363. doi: 10.3389/fpls.2022.907363. eCollection 2022. Front Plant Sci. 2022. PMID: 35812981 Free PMC article. Review.
-
Interspecific reproductive barriers in the tomato clade: opportunities to decipher mechanisms of reproductive isolation.Sex Plant Reprod. 2011 Sep;24(3):171-87. doi: 10.1007/s00497-010-0155-7. Epub 2010 Nov 14. Sex Plant Reprod. 2011. PMID: 21076968 Review.
-
CRISPR/Cas9 Gene Drive: Growing Pains for a New Technology.Genetics. 2017 Mar;205(3):1037-1039. doi: 10.1534/genetics.116.198887. Genetics. 2017. PMID: 28270527 Free PMC article. No abstract available.
-
Fine mapping of the sunflower resistance locus Pl(ARG) introduced from the wild species Helianthus argophyllus.Theor Appl Genet. 2010 Nov;121(8):1633-44. doi: 10.1007/s00122-010-1416-4. Epub 2010 Aug 11. Theor Appl Genet. 2010. PMID: 20700574 Free PMC article.
References
-
- Arnold, M. L., 1993. Iris nelsonii (Iridaceae)—Origin and genetic composition of a homoploid hybrid species. Amer. J. Bot. 80: 577–583. - PubMed
-
- Barton, N., and B. O. Bengtsson, 1986. The barrier to genetic exchange between hybridizing populations. Heredity 57: 357–376. - PubMed
-
- Berry, S. T., A. J. Leon, C. C. Hanfrey, P. Challis and A. Burkholz et al., 1995. Molecular marker analysis of Helianthus annuus L. 2. Construction of an RFLP linkage map for cultivated sunflower. Theor. Appl. Genet. 91: 195–199. - PubMed
-
- Borgen, L., I. Leitch and A. Santos-Guerra, 2003. Genome organization in diploid hybrid species of Argyranthemum (Asteraceae) in the Canary Islands. Bot. J. Linn. Soc. 141: 491–501.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

