A digital atlas to characterize the mouse brain transcriptome
- PMID: 16184189
- PMCID: PMC1215388
- DOI: 10.1371/journal.pcbi.0010041
A digital atlas to characterize the mouse brain transcriptome
Abstract
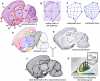
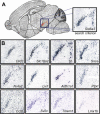
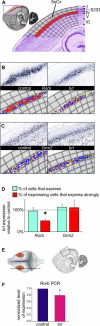
Massive amounts of data are being generated in an effort to represent for the brain the expression of all genes at cellular resolution. Critical to exploiting this effort is the ability to place these data into a common frame of reference. Here we have developed a computational method for annotating gene expression patterns in the context of a digital atlas to facilitate custom user queries and comparisons of this type of data. This procedure has been applied to 200 genes in the postnatal mouse brain. As an illustration of utility, we identify candidate genes that may be related to Parkinson disease by using the expression of a dopamine transporter in the substantia nigra as a search query pattern. In addition, we discover that transcription factor Rorb is down-regulated in the barrelless mutant relative to control mice by quantitative comparison of expression patterns in layer IV somatosensory cortex. The semi-automated annotation method developed here is applicable to a broad spectrum of complex tissues and data modalities.
Conflict of interest statement
Figures

References
-
- Neidhardt L, Gasca S, Wertz K, Obermayr F, Worpenberg S, et al. Large-scale screen for genes controlling mammalian embryogenesis, using high-throughput gene expression analysis in mouse embryos. Mech Dev. 2000;98:77–94. - PubMed
-
- Visel A, Thaller C, Eichele G. GenePaint.org: An atlas of gene expression patterns in the mouse embryo. Nucleic Acids Res. 2004;32:D552–D556. - PMC - PubMed
-
- Boguski MS, Jones AR. Neurogenomics: At the intersection of neurobiology and genome sciences. Nat Neurosci. 2004;7:429–433. - PubMed
-
- Gray PA, Fu H, Luo P, Zhao Q, Yu J, et al. Mouse brain organization revealed through direct genome-scale TF expression analysis. Science. 2004;306:2255–2257. - PubMed
-
- Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, et al. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425:917–925. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

