Dnm1 forms spirals that are structurally tailored to fit mitochondria
- PMID: 16186251
- PMCID: PMC2171542
- DOI: 10.1083/jcb.200506078
Dnm1 forms spirals that are structurally tailored to fit mitochondria
Abstract
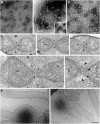
Dynamin-related proteins (DRPs) are large self-assembling GTPases whose common function is to regulate membrane dynamics in a variety of cellular processes. Dnm1, which is a yeast DRP (Drp1/Dlp1 in humans), is required for mitochondrial division, but its mechanism is unknown. We provide evidence that Dnm1 likely functions through self-assembly to drive the membrane constriction event that is associated with mitochondrial division. Two regulatory features of Dnm1 self-assembly were also identified. Dnm1 self-assembly proceeded through a rate-limiting nucleation step, and nucleotide hydrolysis by assembled Dnm1 structures was highly cooperative with respect to GTP. Dnm1 formed extended spirals, which possessed diameters greater than those of dynamin-1 spirals but whose sizes, remarkably, were equal to those of mitochondrial constriction sites in vivo. These data suggest that Dnm1 has evolved to form structures that fit the dimensions of mitochondria.
Figures




References
-
- Chen, Y.J., P. Zhang, E.H. Egelman, and J.E. Hinshaw. 2004. The stalk region of dynamin drives the constriction of dynamin tubes. Nat. Struct. Mol. Biol. 11:574–575. - PubMed
-
- Danino, D., K.H. Moon, and J.E. Hinshaw. 2004. Rapid constriction of lipid bilayers by the mechanochemical enzyme dynamin. J. Struct. Biol. 147:259–267. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

