Human mismatch repair: reconstitution of a nick-directed bidirectional reaction
- PMID: 16188885
- PMCID: PMC1435381
- DOI: 10.1074/jbc.M509701200
Human mismatch repair: reconstitution of a nick-directed bidirectional reaction
Abstract
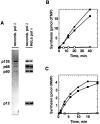
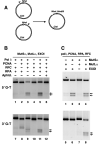
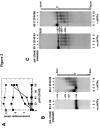
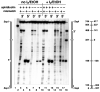
Bidirectional mismatch repair directed by a strand break located 3' or 5' to the mispair has been reconstituted using seven purified human activities: MutSalpha, MutLalpha, EXOI, replication protein A (RPA), proliferating cell nuclear antigen (PCNA), replication factor C (RFC) and DNA polymerase delta. In addition to DNA polymerase delta, PCNA, RFC, and RPA, 5'-directed repair depends on MutSalpha and EXOI, whereas 3'-directed mismatch correction also requires MutLalpha. The repair reaction displays specificity for DNA polymerase delta, an effect that presumably reflects interactions with other repair activities. Because previous studies have suggested potential involvement of the editing function of a replicative polymerase in mismatch-provoked excision, we have evaluated possible participation of DNA polymerase delta in the excision step of repair. RFC and PCNA dramatically activate polymerase delta-mediated hydrolysis of a primer-template. Nevertheless, the contribution of the polymerase to mismatch-provoked excision is very limited, both in the purified system and in HeLa extracts, as judged by in vitro assay using nicked circular heteroplex DNAs. Thus, excision and repair in the purified system containing polymerase delta are reduced 10-fold upon omission of EXOI or by substitution of a catalytically dead form of the exonuclease. Furthermore, aphidicolin inhibits both 3'- and 5'-directed excision in HeLa nuclear extracts by only 20-30%. Although this modest inhibition could be because of nonspecific effects, it may indicate limited dependence of bidirectional excision on an aphidicolin-sensitive DNA polymerase.
Figures

Similar articles
-
Analysis of the excision step in human DNA mismatch repair.Methods Enzymol. 2006;408:273-84. doi: 10.1016/S0076-6879(06)08017-7. Methods Enzymol. 2006. PMID: 16793375
-
A defined human system that supports bidirectional mismatch-provoked excision.Mol Cell. 2004 Jul 2;15(1):31-41. doi: 10.1016/j.molcel.2004.06.016. Mol Cell. 2004. PMID: 15225546
-
Reconstitution of 5'-directed human mismatch repair in a purified system.Cell. 2005 Sep 9;122(5):693-705. doi: 10.1016/j.cell.2005.06.027. Cell. 2005. PMID: 16143102
-
DNA mismatch repair and mutation avoidance pathways.J Cell Physiol. 2002 Apr;191(1):28-41. doi: 10.1002/jcp.10077. J Cell Physiol. 2002. PMID: 11920679 Review.
-
Eukaryotic DNA mismatch repair.Curr Opin Genet Dev. 1999 Feb;9(1):89-96. doi: 10.1016/s0959-437x(99)80013-6. Curr Opin Genet Dev. 1999. PMID: 10072354 Review.
Cited by
-
DNA mismatch repair and Lynch syndrome.J Mol Histol. 2006 Sep;37(5-7):271-83. doi: 10.1007/s10735-006-9038-5. Epub 2006 Jul 4. J Mol Histol. 2006. PMID: 16821093 Review.
-
Orc6 is a component of the replication fork and enables efficient mismatch repair.Proc Natl Acad Sci U S A. 2022 May 31;119(22):e2121406119. doi: 10.1073/pnas.2121406119. Epub 2022 May 27. Proc Natl Acad Sci U S A. 2022. PMID: 35622890 Free PMC article.
-
Identification of regulatory factor X as a novel mismatch repair stimulatory factor.J Biol Chem. 2008 May 9;283(19):12730-5. doi: 10.1074/jbc.M800460200. Epub 2008 Mar 4. J Biol Chem. 2008. PMID: 18319249 Free PMC article.
-
Evolution of the methyl directed mismatch repair system in Escherichia coli.DNA Repair (Amst). 2016 Feb;38:32-41. doi: 10.1016/j.dnarep.2015.11.016. Epub 2015 Dec 2. DNA Repair (Amst). 2016. PMID: 26698649 Free PMC article. Review.
-
Ribonucleotides misincorporated into DNA act as strand-discrimination signals in eukaryotic mismatch repair.Mol Cell. 2013 May 9;50(3):323-32. doi: 10.1016/j.molcel.2013.03.019. Epub 2013 Apr 18. Mol Cell. 2013. PMID: 23603115 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

