ADP-ribose-1"-monophosphatase: a conserved coronavirus enzyme that is dispensable for viral replication in tissue culture
- PMID: 16188975
- PMCID: PMC1235854
- DOI: 10.1128/JVI.79.20.12721-12731.2005
ADP-ribose-1"-monophosphatase: a conserved coronavirus enzyme that is dispensable for viral replication in tissue culture
Abstract
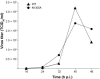
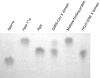
Replication of the approximately 30-kb plus-strand RNA genome of coronaviruses and synthesis of an extensive set of subgenome-length RNAs are mediated by the replicase-transcriptase, a membrane-bound protein complex containing several cellular proteins and up to 16 viral nonstructural proteins (nsps) with multiple enzymatic activities, including protease, polymerase, helicase, methyltransferase, and RNase activities. To get further insight into the replicase gene-encoded functions, we characterized the coronavirus X domain, which is part of nsp3 and has been predicted to be an ADP-ribose-1"-monophosphate (Appr-1"-p) processing enzyme. Bacterially expressed forms of human coronavirus 229E (HCoV-229E) and severe acute respiratory syndrome-coronavirus X domains were shown to dephosphorylate Appr-1"-p, a side product of cellular tRNA splicing, to ADP-ribose in a highly specific manner. The enzyme had no detectable activity on several other nucleoside phosphates. Guided by the crystal structure of AF1521, an X domain homolog from Archaeoglobus fulgidus, potential active-site residues of the HCoV-229E X domain were targeted by site-directed mutagenesis. The data suggest that the HCoV-229E replicase polyprotein residues, Asn 1302, Asn 1305, His 1310, Gly 1312, and Gly 1313, are part of the enzyme's active site. Characterization of an Appr-1"-pase-deficient HCoV-229E mutant revealed no significant effects on viral RNA synthesis and virus titer, and no reversion to the wild-type sequence was observed when the mutant virus was passaged in cell culture. The apparent dispensability of the conserved X domain activity in vitro indicates that coronavirus replicase polyproteins have evolved to include nonessential functions. The biological significance of the novel enzymatic activity in vivo remains to be investigated.
Figures



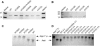

References
-
- Allen, M. D., A. M. Buckle, S. C. Cordell, J. Lowe, and M. Bycroft. 2003. The crystal structure of AF1521 a protein from Archaeoglobus fulgidus with homology to the non-histone domain of macroH2A. J. Mol. Biol. 330:503-511. - PubMed
-
- Anand, K., J. Ziebuhr, P. Wadhwani, J. R. Mesters, and R. Hilgenfeld. 2003. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science 300:1763-1767. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

