Site-directed mutagenesis of large DNA palindromes: construction and in vitro characterization of herpes simplex virus type 1 mutants containing point mutations that eliminate the oriL or oriS initiation function
- PMID: 16188981
- PMCID: PMC1235857
- DOI: 10.1128/JVI.79.20.12783-12797.2005
Site-directed mutagenesis of large DNA palindromes: construction and in vitro characterization of herpes simplex virus type 1 mutants containing point mutations that eliminate the oriL or oriS initiation function
Abstract
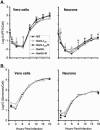
Technical challenges associated with mutagenesis of the large oriL palindrome have hindered comparisons of the functional roles of the herpes simplex virus type 1 (HSV-1) origins of DNA replication, oriL and oriS, in viral replication and pathogenesis. To address this problem, we have developed a novel PCR-based strategy to introduce site-specific mutations into oriL and other large palindromes. Using this strategy, we generated three plasmids containing mutant forms of oriL, i.e., pDoriL-I(L), pDoriL-I(R), and pDoriL-I(LR), containing point mutations in the left, right, and both copies, respectively, of the origin binding protein (OBP) binding site (site I) which eliminate OBP binding. In in vitro DNA replication assays, plasmids with mutations in only one arm of the palindrome supported origin-dependent DNA replication, whereas plasmids with symmetrical mutations in both arms of the palindrome were replication incompetent. An analysis of the cloned mutant plasmids used in replication assays revealed that a fraction of each plasmid mutated in only one arm of the palindrome had lost the site I mutation. In contrast, plasmids containing symmetrical mutations in both copies of site I retained both mutations. These observations demonstrate that the single site I mutations in pDoriL-I(L) and pDoriL-I(R) are unstable upon propagation in bacteria and suggest that functional forms of both the left and right copies of site I are required to initiate DNA replication at oriL. To examine the role of oriL and oriS site I in virus replication, we introduced the two site I mutations in pDoriL-I(LR) into HSV-1 DNA to yield the mutant virus DoriL-I(LR) and the same point mutations into the single site I sequence present in both copies of oriS to yield the mutant virus DoriS-I. In Vero cells and primary rat embryonic cortical neurons (PRN) infected with either mutant virus, viral DNA synthesis and viral replication were efficient, confirming that the two origins can substitute functionally for one another in vitro. Measurement of the levels of oriL and oriS flanking gene transcripts revealed a modest alteration in the kinetics of ICP8 transcript accumulation in DoriL-I(LR)-infected PRN, but not in Vero cells, implicating a cell-type-specific role for oriL in regulating ICP8 transcription.
Figures




References
-
- Adom, J. N., F. Gouilleux, and H. Richard-Foy. 1992. Interaction with the nuclear matrix of a chimeric construct containing a replication origin and a transcription unit. Biochim. Biophys. Acta 1171:187-197. - PubMed
-
- Baker, R. O., L. B. Murata, M. S. Dodson, and J. D. Hall. 2000. Purification and characterization of OF-1, a host factor implicated in herpes simplex replication. J. Biol. Chem. 275:30050-30057. - PubMed
-
- Bode, J., C. Benham, A. Knopp, and C. Mielke. 2000. Transcriptional augmentation: modulation of gene expression by scaffold/matrix-attached regions (S/MAR elements). Crit. Rev. Eukaryot. Gene Expr. 10:73-90. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

