Structural genomics of the severe acute respiratory syndrome coronavirus: nuclear magnetic resonance structure of the protein nsP7
- PMID: 16188992
- PMCID: PMC1235862
- DOI: 10.1128/JVI.79.20.12905-12913.2005
Structural genomics of the severe acute respiratory syndrome coronavirus: nuclear magnetic resonance structure of the protein nsP7
Abstract
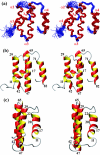
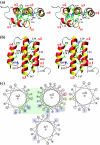
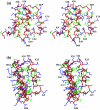
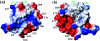
Here, we report the three-dimensional structure of severe acute respiratory syndrome coronavirus (SARS-CoV) nsP7, a component of the SARS-CoV replicase polyprotein. The coronavirus replicase carries out regulatory tasks involved in the maintenance, transcription, and replication of the coronavirus genome. nsP7 was found to assume a compact architecture in solution, which is comprised primarily of helical secondary structures. Three helices (alpha2 to alpha4) form a flat up-down-up antiparallel alpha-helix sheet. The N-terminal segment of residues 1 to 22, containing two turns of alpha-helix and one turn of 3(10)-helix, is packed across the surface of alpha2 and alpha3 in the helix sheet, with the alpha-helical region oriented at a 60 degrees angle relative to alpha2 and alpha3. The surface charge distribution is pronouncedly asymmetrical, with the flat surface of the helical sheet showing a large negatively charged region adjacent to a large hydrophobic patch and the opposite side containing a positively charged groove that extends along the helix alpha1. Each of these three areas is thus implicated as a potential site for protein-protein interactions.
Figures

References
-
- Bowie, A. G., J. Zhan, and W. L. Marshall. 2004. Viral appropriation of apoptotic and NF-κB signaling pathways. J. Cell. Biochem. 91:1099-1108. - PubMed
-
- Cornell, W. D., P. Cieplak, C. I. Bayly, I. R. Gould, K. M. Merz, Jr., D. M. Ferguson, D. C. Spellmeyer, T. Fox, J. W. Caldwell, and P. A. Kollman. 1995. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 117:5179-5197.
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

