A role for herpesvirus entry mediator as the receptor for herpes simplex virus 1 entry into primary human trabecular meshwork cells
- PMID: 16189018
- PMCID: PMC1235852
- DOI: 10.1128/JVI.79.20.13173-13179.2005
A role for herpesvirus entry mediator as the receptor for herpes simplex virus 1 entry into primary human trabecular meshwork cells
Abstract
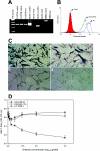
The human eye is an important target for infection with herpes simplex virus 1 (HSV-1). Damage to cells forming the trabeculum of the eye by HSV-1 infection could contribute to the development of glaucoma, a major blinding disease. Primary cultures of human trabecular meshwork cells were used as an in vitro model to demonstrate the ability of HSV-1 to enter into and establish a productive infection of the trabeculum. Blocking of entry by anti-herpesvirus entry mediator (HVEM) antibody implicated HVEM as the major receptor for HSV-1 infection.
Figures


References
-
- Amano, S., T. Oshika, Y. Kaji, J. Numaga, M. Matsubara, and M. Araie. 1999. Herpes simplex virus in the trabeculum of an eye with corneal endotheliitis. Am. J. Ophthalmol. 127:721-722. - PubMed
-
- Bustos, D. E., and S. S. Atherton. 2002. Detection of herpes simplex virus type-1 in human ciliary ganglia. Investig. Ophthalmol. Vis. Sci. 43:2244-2249. - PubMed
-
- Cocchi, F., L. Menotti, P. Mirandola, M. Lopez, and G. Campadelli-Fiume. 1998. The ectodomain of a novel member of the immunoglobulin subfamily related to the poliovirus receptor has the attributes of a bona fide receptor for herpes simplex virus types 1 and 2 in human cells. J. Virol. 72:9992-10002. - PMC - PubMed
-
- Eberle, F., P. Dubreuil, M. G. Mattei, E. Devilard, and M. Lopez. 1995. The human PRR2 gene, related to the human poliovirus receptor gene (PVR), is the true homolog of the murine MPH gene. Gene 159:267-272. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

