Characterization of the naturally occurring oxacillinase of Acinetobacter baumannii
- PMID: 16189095
- PMCID: PMC1251506
- DOI: 10.1128/AAC.49.10.4174-4179.2005
Characterization of the naturally occurring oxacillinase of Acinetobacter baumannii
Abstract
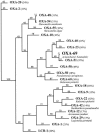
A chromosomally encoded oxacillinase, OXA-69, was characterized from Acinetobacter baumannii AYE. beta-Lactamase OXA-69 shared 97% amino acid identity with the recently described OXA-51 enzyme of A. baumannii and 62 and 56% amino acid identity with the carbapenem-hydrolyzing oxacillinases OXA-24 and OXA-23, respectively. Biochemical characterization of the purified OXA-69 revealed a narrow-spectrum hydrolysis profile but including, at a low level, imipenem and meropenem. By PCR and sequencing bla(OXA-69)-like genes were identified in all A. baumannii strains tested (n = 12), suggesting that this oxacillinase is naturally occurring in that species.
Figures
References
-
- Barbe, V., D. Vallenet, N. Fonknechten, A. Kreimeyer, S. Oztas, L. Labarre, S. Cruveiller, C. Robert, S. Duprat, P. Wincker, L. N. Ornston, J. Weissenbach, P. Marliere, G. N. Cohen, and C. Medigue. 2004. Unique features revealed by the genome sequence of Acinetobacter sp. ADP1, a versatile and naturally transformation competent bacterium. Nucleic Acids Res. 32:5766-5779. - PMC - PubMed
-
- Bergogne-Bérézin, E., M. L. Joly-Guillou, and K. J. Towner. 1996. Epidemiology of Acinetobacter spp. p. 71-100. In K. J. Towner et al. (ed.), Acinetobacter: microbiology, epidemiology, infections, management. CRC Press, Inc., Boca Raton, Fla.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

