Macrolide efflux in Streptococcus pneumoniae is mediated by a dual efflux pump (mel and mef) and is erythromycin inducible
- PMID: 16189099
- PMCID: PMC1251515
- DOI: 10.1128/AAC.49.10.4203-4209.2005
Macrolide efflux in Streptococcus pneumoniae is mediated by a dual efflux pump (mel and mef) and is erythromycin inducible
Abstract
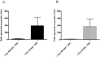
Macrolide resistance in Streptococcus pneumoniae due to efflux has emerged as an important worldwide clinical problem over the past decade. Efflux is mediated by the genes of the genetic element mega (macrolide efflux genetic assembly) and related elements, such as Tn1207.1. These elements contain two adjacent genes, mef (mefE or mefA) and the closely related mel gene (msrA homolog), encoding a proton motive force pump and a putative ATP-binding cassette transporter homolog, and are transcribed as an operon (M. Del Grosso et al., J. Clin. Microbiol. 40:774-778, 2004; K. Gay and D. S. Stephens, J. Infect. Dis. 184:56-65, 2001; and M. Santagati et al., Antimicrob. Agents Chemother. 44:2585-2587, 2000). Previous studies have shown that Mef is required for macrolide resistance in S. pneumoniae; however, the contribution of Mel has not been fully determined. Independent deletions were constructed in mefE and mel in the serotype 14 macrolide-resistant strains GA16638 (erythromycin [Em] MIC, 8 to 16 microg/ml) and GA17719 (Em MIC, 2 to 4 microg/ml), which contain allelic variations in the mega element. The MICs to erythromycin were significantly reduced for the independent deletion mutants of both mefE and mel compared to those of the parent strains and further reduced threefold to fourfold to Em MICs of <0.15 microg/ml with mefE mel double mutants. Using quantitative reverse transcription-PCR, the expression of mefE in the mel deletion mutants was increased more than 10-fold. However, in the mefE deletion mutants, the expression of mel did not differ significantly from the parent strains. The expression of both mefE and mel was inducible by erythromycin. These data indicate a requirement for both Mef and Mel in the novel efflux-mediated macrolide resistance system in S. pneumoniae and other gram-positive bacteria and that the system is inducible by macrolides.
Figures

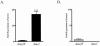

References
-
- Banks, D. J., S. F. Porcella, K. D. Barbian, J. M. Martin, and J. M. Musser. 2003. Structure and distribution of an unusual chimeric genetic element encoding macrolide resistance in phylogenetically diverse clones of group A streptococcus. J. Infect. Dis. 188:1898-1908. - PubMed
-
- Canton, R., A. Mazzariol, M. I. Morosini, F. Baquero, and G. Cornaglia. 2005. Telithromycin activity is reduced by efflux in Streptococcus pyogenes. J. Antimicrob. Chemother. 55:489-495. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

