Mutations conferring resistance to a hepatitis C virus (HCV) RNA-dependent RNA polymerase inhibitor alone or in combination with an HCV serine protease inhibitor in vitro
- PMID: 16189112
- PMCID: PMC1251566
- DOI: 10.1128/AAC.49.10.4305-4314.2005
Mutations conferring resistance to a hepatitis C virus (HCV) RNA-dependent RNA polymerase inhibitor alone or in combination with an HCV serine protease inhibitor in vitro
Abstract
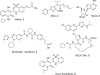
Compounds A-782759 (an N-1-aza-4-hydroxyquinolone benzothiadiazine) and BILN-2061 are specific anti-hepatitis C virus (HCV) agents that inhibit the RNA-dependent RNA polymerase and the NS3 serine protease, respectively. Both compounds display potent activity against HCV replicons in tissue culture. In order to characterize the development of resistance to these anti-HCV agents, HCV subgenomic 1b-N replicon cells were cultured with A-782759 alone or in combination with BILN-2061 at concentrations 10 times above their corresponding 50% inhibitory concentrations in the presence of neomycin. Single substitutions in the NS5B polymerase gene (H95Q, N411S, M414L, M414T, or Y448H) resulted in substantial decreases in susceptibility to A-782759. Similarly, replicons containing mutations in the NS5B polymerase gene (M414L or M414T), together with single mutations in the NS3 protease gene (A156V or D168V), conferred high levels of resistance to both A-782759 and BILN-2061. However, the A-782759-resistant mutants remained susceptible to nucleoside and two other classes of nonnucleoside NS5B polymerase inhibitors, as well as interferon. In addition, we found that the frequency of replicons resistant to both compounds was significantly lower than the frequency of resistance to the single compound. Furthermore, the dually resistant mutants displayed significantly reduced replication capacities compared to the wild-type replicon. These findings provide strategic guidance for the future treatment of HCV infection.
Figures
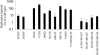
References
-
- Bartenschlager, R. 1999. The NS3/4A proteinase of the hepatitis C virus: unravelling structure and function of an unusual enzyme and a prime target for antiviral therapy. J. Viral Hepat. 6:165-181. - PubMed
-
- Beaulieu, P. 2004. Allosteric inhibitors of the hepatitis C virus NS5B polymerase: towards novel anti-HCV therapies, abstr. 1. Abstr. 29th Nat. Med. Chem. Symp. American Chemical Society and University of Wisconsin—Madison, Madison, Wis.
-
- Carroll, S. S., J. E. Tomassini, M. Bosserman, K. Getty, M. W. Stahlhut, A. B. Eldrup, B. Bhat, D. Hall, A. L. Simcoe, R. LaFemina, C. A. Rutkowski, B. Wolanski, Z. Yang, G. Migliaccio, R. De Francesco, L. C. Kuo, M. MacCoss, and D. B. Olsen. 2003. Inhibition of hepatitis C virus RNA replication by 2′-modified nucleoside analogs. J. Biol. Chem. 278:11979-11984. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

