No common denominator for breast cancer lymph node metastasis
- PMID: 16189523
- PMCID: PMC2361648
- DOI: 10.1038/sj.bjc.6602794
No common denominator for breast cancer lymph node metastasis
Abstract
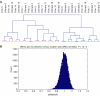
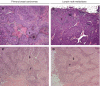
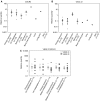
The axillary lymph node status is the most powerful prognostic factor for breast cancer patients to date. The molecular mechanisms that control lymph node metastasis, however, remain poorly understood. To define patterns of genes or gene regulatory pathways that drive breast cancer lymph node metastasis, we compared the gene expression profiles of 15 primary breast carcinomas and their matching lymph node metastases using microarrays. In general, primary breast carcinomas and lymph node metastases do not differ at the transcriptional level by a common subset of genes. No classifier or single gene discriminating the group of primary tumours from those of the lymph node metastases could be identified. Also, in a series of 295 breast tumours, no classifier predicting lymph node metastasis could be developed. However, subtle differences in the expression of genes involved in extracellular-matrix organisation and growth factor signalling are detected in individual pairs of matching primary and metastatic tumours. Surprisingly, however, different sets of these genes are either up- or downregulated in lymph node metastases. Our data suggest that breast carcinomas do not use a shared gene set to accomplish lymph node metastasis.
Figures

References
-
- Al-Shahrour F, Diaz-Uriarte R, Dopazo J (2004) FatiGO: a web tool for finding significant associations of Gene Ontology terms with groups of genes. Bioinformatics 20: 578–580 - PubMed
-
- Balkwill F (2004) Cancer and the chemokine network. Nat Rev Cancer 4: 540–550 - PubMed
-
- Bono P, Wasenius V, Heikkila P, Lundin J, Jackson D, Joensuu H (2004) High LYVE-1-positive lymphatic vessel numbers are associated with poor outcome in breast cancer. Clin Cancer Res 10: 7144–7149 - PubMed
-
- Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC, Gaasterland T, Glenisson P, Holstege FC, Kim IF, Markowitz V, Matese JC, Parkinson H, Robinson A, Sarkans U, Schulze-Kremer S, Stewart J, Taylor R, Vilo J, Vingron M (2001) Minimum information about a microarray experiment (MIAME) – toward standards for microarray data. Nat Genet 29: 365–371 - PubMed
-
- Chambers AF, Groom AC, MacDonald IC (2002) Dissemination and growth of cancer cells in metastatic sites. Nat Rev Cancer 2: 563–572 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

