Wrapping things up about virus RNA replication
- PMID: 16190978
- PMCID: PMC7169867
- DOI: 10.1111/j.1600-0854.2005.00339.x
Wrapping things up about virus RNA replication
Abstract
All single-stranded 'positive-sense' RNA viruses that infect mammalian, insect or plant cells rearrange internal cellular membranes to provide an environment facilitating virus replication. A striking feature of these unique membrane structures is the induction of 70-100 nm vesicles (either free within the cytoplasm, associated with other induced vesicles or bound within a surrounding membrane) harbouring the viral replication complex (RC). Although similar in appearance, the cellular composition of these vesicles appears to vary for different viruses, implying different organelle origins for the intracellular sites of viral RNA replication. Genetic analysis has revealed that induction of these membrane structures can be attributed to a particular viral gene product, usually a non-structural protein. This review will highlight our current knowledge of the formation and composition of virus RCs and describe some of the similarities and differences in RNA-membrane interactions observed between the virus families Flaviviridae and Picornaviridae.
Figures
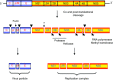
 depict cleavage by the viral‐encoded protease NS3 with cofactor NS2B. The cleavage between NS1 and NS2A is currently not well understood but is performed by a host‐cell protease in the lumen of the endoplasmic recticulum.
depict cleavage by the viral‐encoded protease NS3 with cofactor NS2B. The cleavage between NS1 and NS2A is currently not well understood but is performed by a host‐cell protease in the lumen of the endoplasmic recticulum.


References
-
- Bienz K, Egger D, Pasamontes L. Association of polioviral proteins of the P2 genomic region with the viral replication complex and virus‐induced membrane synthesis as visualized by electron microscopic immunocytochemistry and autoradiography. Virology 1987;160: 220–226. - PubMed
-
- Magliano D, Marshall JA, Bowden DS, Vardaxis N, Meanger J, Lee JY. Rubella virus replication complexes are virus‐modified lysosomes. Virology 1998;240: 57–63. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

