Optimized between-group classification: a new jackknife-based gene selection procedure for genome-wide expression data
- PMID: 16191195
- PMCID: PMC1261161
- DOI: 10.1186/1471-2105-6-239
Optimized between-group classification: a new jackknife-based gene selection procedure for genome-wide expression data
Abstract
Background: A recent publication described a supervised classification method for microarray data: Between Group Analysis (BGA). This method which is based on performing multivariate ordination of groups proved to be very efficient for both classification of samples into pre-defined groups and disease class prediction of new unknown samples. Classification and prediction with BGA are classically performed using the whole set of genes and no variable selection is required. We hypothesize that an optimized selection of highly discriminating genes might improve the prediction power of BGA.
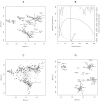
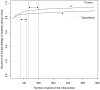
Results: We propose an optimized between-group classification (OBC) which uses a jackknife-based gene selection procedure. OBC emphasizes classification accuracy rather than feature selection. OBC is a backward optimization procedure that maximizes the percentage of between group inertia by removing the least influential genes one by one from the analysis. This selects a subset of highly discriminative genes which optimize disease class prediction. We apply OBC to four datasets and compared it to other classification methods.
Conclusion: OBC considerably improved the classification and predictive accuracy of BGA, when assessed using independent data sets and leave-one-out cross-validation.
Availability: The R code is freely available [see Additional file 1] as well as supplementary information [see Additional file 2].
Figures



Similar articles
-
A new method for class prediction based on signed-rank algorithms applied to Affymetrix microarray experiments.BMC Bioinformatics. 2008 Jan 11;9:16. doi: 10.1186/1471-2105-9-16. BMC Bioinformatics. 2008. PMID: 18190711 Free PMC article.
-
GeneSrF and varSelRF: a web-based tool and R package for gene selection and classification using random forest.BMC Bioinformatics. 2007 Sep 3;8:328. doi: 10.1186/1471-2105-8-328. BMC Bioinformatics. 2007. PMID: 17767709 Free PMC article.
-
Bias in error estimation when using cross-validation for model selection.BMC Bioinformatics. 2006 Feb 23;7:91. doi: 10.1186/1471-2105-7-91. BMC Bioinformatics. 2006. PMID: 16504092 Free PMC article.
-
Classification based upon gene expression data: bias and precision of error rates.Bioinformatics. 2007 Jun 1;23(11):1363-70. doi: 10.1093/bioinformatics/btm117. Epub 2007 Mar 28. Bioinformatics. 2007. PMID: 17392326 Review.
-
Advances in metaheuristics for gene selection and classification of microarray data.Brief Bioinform. 2010 Jan;11(1):127-41. doi: 10.1093/bib/bbp035. Epub 2009 Sep 29. Brief Bioinform. 2010. PMID: 19789265 Review.
Cited by
-
Stability of gene contributions and identification of outliers in multivariate analysis of microarray data.BMC Bioinformatics. 2008 Jun 20;9:289. doi: 10.1186/1471-2105-9-289. BMC Bioinformatics. 2008. PMID: 18570644 Free PMC article.
-
Expression profiling in granulomatous lung disease.Proc Am Thorac Soc. 2007 Jan;4(1):101-7. doi: 10.1513/pats.200607-140JG. Proc Am Thorac Soc. 2007. PMID: 17202298 Free PMC article. Review.
References
-
- Li L, Pedersen LG, Darden TA, Weinberg CR. Class prediction and discovery based on gene expression data. Genome Information Systems and Technology. 2001.
-
- Xiong M, Jin L, Li W, Boerwinkle E. Computational methods for gene expression-based tumor classification. Biotechniques. 2000;29:1264–8. 1270. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

