Characterization of the bisintercalative DNA binding mode of a bifunctional platinum-acridine agent
- PMID: 16192574
- PMCID: PMC1236979
- DOI: 10.1093/nar/gki869
Characterization of the bisintercalative DNA binding mode of a bifunctional platinum-acridine agent
Abstract
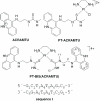
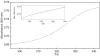
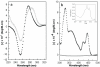
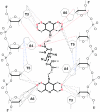
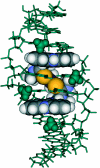
The DNA interactions of PT-BIS(ACRAMTU) ([Pt(en)(ACRAMTU)2](NO3)4; ACRAMTU = 1-[2-(acridin-9-ylamino)ethyl]-1,3-dimethylthiourea, en = ethylenediamine), a bifunctional platinum-acridine conjugate, have been studied in native and synthetic double-stranded DNAs and model duplexes using various biophysical techniques. These include ethidium-DNA fluorescence quenching and thermal melting experiments, circular dichroism (CD) spectroscopy and plasmid unwinding assays. In addition, the binding mode was studied in a short octamer by NMR spectroscopy in conjunction with molecular modeling. In alternating copolymers, PT-BIS(ACRAMTU) shows a distinct preference for poly(dA-dT)2, which is approximately 3-fold higher than that of ACRAMTU. In the ligand-oligomer complex, d(GCTATAGC)2.PT-BIS(ACRAMTU) (complex I*), PT-BIS(ACRAMTU) increases the thermal stability of the B-form host duplex by DeltaT(m) > 30 K (CD and UV melting experiments). The agent unwinds pSP73 plasmid DNA by 44(+/-2) degrees per bound molecule, indicating bisintercalative binding. A 2-D NMR study unequivocally demonstrates that PT-BIS(ACRAMTU)'s chromophores deeply bisintercalate into the 5'-TA/TA base pair steps in I*, while the platinum linker lies in the minor groove. An AMBER model reflecting the NMR results shows that bracketing of the central AT base pairs in a classical nearest neighbor excluded fashion is feasible. PT-BIS(ACRAMTU) inhibits DNA hydrolysis by BstZ17 I at the enzyme's restriction site, GTA downward arrowTAC. Possible consequences for other relevant DNA-protein interactions, such as those involved in TATA-box-mediated transcription initiation and the utility of the platinum-intercalator technology for the design of sequence-specific agents are discussed.
Figures




References
-
- Chaires J.B. Drug–DNA interactions. Curr. Opin. Struct. Biol. 1998;8:314–320. - PubMed
-
- Gago F. Stacking interactions and intercalative DNA binding. Methods. 1998;14:277–292. - PubMed
-
- Foster B.J., Clagett-Carr K., Shoemaker D.D., Suffness M., Plowman J., Trissel L.A., Grieshaber C.K., Leyland-Jones B. Echinomycin: the first bifunctional intercalating agent in clinical trials. Invest. New Drugs. 1985;3:403–410. - PubMed
-
- Leroy J.L., Gao X.L., Misra V., Gueron M., Patel D.J. Proton exchange in DNA-luzopeptin and DNA-echinomycin bisintercalation complexes: rates and processes of base-pair opening. Biochemistry. 1992;31:1407–1415. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

