Determination of complex isotopomer patterns in isotopically labeled compounds by mass spectrometry
- PMID: 16194110
- PMCID: PMC2268020
- DOI: 10.1021/ac0509354
Determination of complex isotopomer patterns in isotopically labeled compounds by mass spectrometry
Abstract
A classic problem in analytical chemistry has been determination of individual components in a mixture without availability of the pure individual components. Measurement of the distribution of isotopomers in a labeled compound or mixture of labeled compounds is an example of this problem that is commonly encountered when stable isotopically labeled metabolites are used to determine in vivo kinetics and metabolism. We present a method that uses the measured mass spectral data of the unlabeled material to represent any and all combinations of isotopomer variations of that material and to determine abundances of these isotopomers. Although examples of the method are presented for gas chromatography/mass spectrometry, the method is applicable to any type of mass spectrometry data. The method also accounts for errors induced by mass spectrometer ionization and resolution effects. To demonstrate this method, we determined the isotopomer distributions of samples of 13C-labeled leucine and glucose for both highly enriched isotopomers and labeled isotopomers present in low abundance against a natural isotopic abundance background. The method accurately and precisely determined isotopomer identity and abundance in the labeled materials without adding noise or error that was not inherent in the original mass spectral data. In examples shown here, isotopomer uncertainties were calculated with relative standard errors of <1% from good quality mass spectral data.
Figures
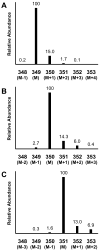

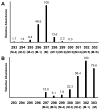
References
-
- Biemann K. Mass Spectrometry: Organic Chemical Applications. McGraw-Hill; New York: 1962.
-
- Tserng KY, Kalhan SC. Am J Physiol Endocrinol Metab. 1983;245:E308–E311. - PubMed
-
- Rosenblatt J, Chinkes D, Wolfe M, Wolfe RR. Am J Physiol Endocrinol Metab. 1992;263:E584–E596. - PubMed
-
- Lee WNP, Whiting JS, Fymat AL, Boettger HG. Biol Mass Spectrom. 1983;10:641–45.
-
- Lee WNP. J Biol Chem. 1989;264:13002–04.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

