The frequency and rate of pilin antigenic variation in Neisseria gonorrhoeae
- PMID: 16194236
- PMCID: PMC2657081
- DOI: 10.1111/j.1365-2958.2005.04838.x
The frequency and rate of pilin antigenic variation in Neisseria gonorrhoeae
Abstract
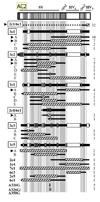
The pilin antigenic variation (Av) system of Neisseria gonorrhoeae (Gc) mediates unidirectional DNA recombination from silent gene copies into the pilin expression locus. A DNA sequencing assay was developed to accurately measure pilin Av in a population of Gc strain FA1090 arising from a defined pilin progenitor under non-selective culture conditions. This assay employs a piliated parental Gc variant with a recA allele whose promoter is replaced by lac-regulatory elements, allowing for controlled induction of pilin Av. From this assay, the frequency of pilin Av was measured as 0.13 recombination events per cell, with a corresponding rate of pilin Av of 4x10(-3) events per cell per generation. Most pilin variants retained the parental piliation phenotype, providing the first comprehensive analysis of piliated variants arising from a piliated progenitor. Sequence analysis of pilin variants revealed that a subset of possible recombination events predominated, which differed between piliated and non-piliated progeny. Pilin Av exhibits the highest reported frequency of any pathogenic gene conversion system and can account for the extensive pilin variation detected during human infection.
Figures


References
-
- Cohen MS, Cannon JG. Human experimentation with Neisseria gonorrhoeae: progress and goals. J Infect Dis. 1999;179 Suppl 2:S375–S379. - PubMed
-
- Haas R, Meyer TF. The repertoire of silent pilus genes in Neisseria gonorrhoeae: evidence for gene conversion. Cell. 1986;44:107–115. - PubMed
-
- Haas R, Veit S, Meyer TF. Silent pilin genes of Neisseria gonorrhoeae MS11 and the occurrence of related hypervariant sequences among other gonococcal isolates. Mol Microbiol. 1992;6:197–208. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

