Induction of dedifferentiation, genomewide transcriptional programming, and epigenetic reprogramming by extracts of carcinoma and embryonic stem cells
- PMID: 16195347
- PMCID: PMC1289416
- DOI: 10.1091/mbc.e05-06-0572
Induction of dedifferentiation, genomewide transcriptional programming, and epigenetic reprogramming by extracts of carcinoma and embryonic stem cells
Abstract
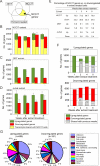
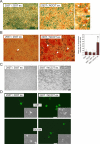
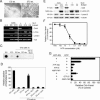
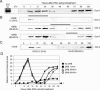
Functional reprogramming of a differentiated cell toward pluripotency may have long-term applications in regenerative medicine. We report the induction of dedifferentiation, associated with genomewide programming of gene expression and epigenetic reprogramming of an embryonic gene, in epithelial 293T cells treated with an extract of undifferentiated human NCCIT carcinoma cells. 293T cells exposed for 1 h to extract of NCCIT cells, but not of 293T or Jurkat T-cells, form defined colonies that are maintained for at least 23 passages in culture. Microarray and quantitative analyses of gene expression reveal that the transition from a 293T to a pluripotent cell phenotype involves a dynamic up-regulation of hundreds of NCCIT genes, concomitant with down-regulation of 293T genes and of indicators of differentiation such as A-type lamins. Up-regulated genes encompass embryonic and stem cell markers, including OCT4, SOX2, NANOG, and Oct4-responsive genes. OCT4 activation is associated with DNA demethylation in the OCT4 promoter and nuclear targeting of Oct4 protein. In fibroblasts exposed to extract of mouse embryonic stem cells, Oct4 activation is biphasic and RNA-PolII dependent, with the first transient rise of Oct4 up-regulation being necessary for the second, long-term activation of Oct4. Genes characteristic of multilineage differentiation potential are also up-regulated in NCCIT extract-treated cells, suggesting the establishment of "multilineage priming." Retinoic acid triggers Oct4 down-regulation, de novo activation of A-type lamins, and nestin. Furthermore, the cells can be induced to differentiate toward neurogenic, adipogenic, osteogenic, and endothelial lineages. The data provide a proof-of-concept that an extract of undifferentiated carcinoma cells can elicit differentiation plasticity in an otherwise more developmentally restricted cell type.
Figures








References
-
- Akashi, K., He, X., Chen, J., Iwasaki, H., Niu, C., Steenhard, B., Zhang, J., Haug, J., and Li, L. (2003). Transcriptional accessibility for genes of multiple tissues and hematopoietic lineages is hierarchically controlled during early hematopoiesis. Blood 101, 383-389. - PubMed
-
- Boquest, A. C., Shahdadfar, A., Fronsdal, K., Sigurjonsson, O., Tunheim, S. H., Collas, P., and Brinchmann, J. E. (2005). Isolation and transcription profiling of purified uncultured human stromal stem cells: alteration of gene expression following in vitro cell culture. Mol. Biol. Cell 16, 1131-1141. - PMC - PubMed
-
- Byrne, J. A., Simonsson, S., Western, P. S., and Gurdon, J. B. (2003). Nuclei of adult mammalian somatic cells are directly reprogrammed to oct-4 stem cell gene expression by amphibian oocytes. Curr. Biol. 13, 1206-1213. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

