A novel motif governs APC-dependent degradation of Drosophila ORC1 in vivo
- PMID: 16195415
- PMCID: PMC1257400
- DOI: 10.1101/gad.1361905
A novel motif governs APC-dependent degradation of Drosophila ORC1 in vivo
Abstract
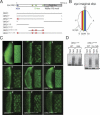
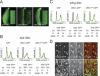
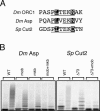
Regulated degradation plays a key role in setting the level of many factors that govern cell cycle progression. In Drosophila, the largest subunit of the origin recognition complex protein 1 (ORC1) is degraded at the end of M phase and throughout much of G1 by anaphase-promoting complexes (APC) activated by Fzr/Cdh1. We show here that none of the previously identified APC motifs targets ORC1 for degradation. Instead, a novel sequence, the O-box, is necessary and sufficient to direct Fzr/Cdh1-dependent polyubiquitylation in vitro and degradation in vivo. The O-box is similar to but distinct from the well characterized D-box. Finally, we show that O-box motifs in two other proteins, Drosophila Abnormal Spindle and Schizosaccharomyces pombe Cut2, contribute to Cdh1-dependent polyubiquitylation in vitro, suggesting that the O-box may mediate degradation of a variety of cell cycle factors.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
