Histone H3 tail positioning and acetylation by the c-Myb but not the v-Myb DNA-binding SANT domain
- PMID: 16195416
- PMCID: PMC1257399
- DOI: 10.1101/gad.355405
Histone H3 tail positioning and acetylation by the c-Myb but not the v-Myb DNA-binding SANT domain
Abstract
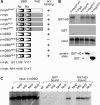
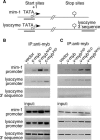
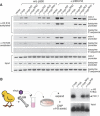
The c-Myb transcription factor coordinates proliferation and differentiation of hematopoietic precursor cells. Myb has three consecutive N-terminal SANT-type repeat domains (R1, R2, R3), two of which (R2, R3) form the DNA-binding domain (DBD). Three amino acid substitutions in R2 alter the way Myb regulates genes and determine the leukemogenicity of the retrovirally transduced v-Myb oncogene. The molecular mechanism of how these mutations unleash the leukemogenic potential of Myb is unknown. Here we demonstrate that the c-Myb-DBD binds to the N-terminal histone tails of H3 and H3.3. C-Myb binding facilitates histone tail acetylation, which is mandatory during activation of prevalent differentiation genes in conjunction with CCAAT enhancer-binding proteins (C/EBP). Leukemogenic mutations in v-Myb eliminate the interaction with H3 and acetylation of H3 tails and abolish activation of endogenous differentiation genes. In primary v-myb-transformed myeloblasts, pharmacologic enhancement of H3 acetylation restored activation of differentiation genes and induced cell differentiation. Our data link a novel chromatin function of c-Myb with lineage-specific expression of differentiation genes and relate the loss of this function with the leukemic conversion of Myb.
Figures




Similar articles
-
Identification of a Myb-responsive enhancer of the chicken C/EBPbeta gene.Oncogene. 2004 Jul 29;23(34):5807-14. doi: 10.1038/sj.onc.1207722. Oncogene. 2004. PMID: 15195136
-
Overexpression of the c-Myb but not its leukemogenic mutant DNA-binding domain increased adipogenic differentiation in mesenchymal stem cells.Biochem Biophys Res Commun. 2011 Apr 1;407(1):202-6. doi: 10.1016/j.bbrc.2011.02.140. Epub 2011 Mar 3. Biochem Biophys Res Commun. 2011. PMID: 21376705
-
tom-1, a novel v-Myb target gene expressed in AMV- and E26-transformed myelomonocytic cells.EMBO J. 1997 Mar 17;16(6):1371-80. doi: 10.1093/emboj/16.6.1371. EMBO J. 1997. PMID: 9135152 Free PMC article.
-
The myb oncogene.Gene Amplif Anal. 1986;4:73-98. Gene Amplif Anal. 1986. PMID: 3333362 Review.
-
Epigenetic control of ovarian function: the emerging role of histone modifications.Mol Cell Endocrinol. 2005 Nov 24;243(1-2):12-8. doi: 10.1016/j.mce.2005.09.005. Epub 2005 Oct 10. Mol Cell Endocrinol. 2005. PMID: 16219412 Review.
Cited by
-
The Myb/SANT domain of the telomere-binding protein TRF2 alters chromatin structure.Nucleic Acids Res. 2009 Aug;37(15):5019-31. doi: 10.1093/nar/gkp515. Epub 2009 Jun 16. Nucleic Acids Res. 2009. PMID: 19531742 Free PMC article.
-
Myb proteins: angels and demons in normal and transformed cells.Front Biosci (Landmark Ed). 2011 Jan 1;16(3):1109-31. doi: 10.2741/3738. Front Biosci (Landmark Ed). 2011. PMID: 21196221 Free PMC article. Review.
-
Lsd1 interacts with cMyb to demethylate repressive histone marks and maintain inner ear progenitor identity.Development. 2018 Feb 21;145(4):dev160325. doi: 10.1242/dev.160325. Development. 2018. PMID: 29437831 Free PMC article.
-
Histone chaperone Jun dimerization protein 2 (JDP2): role in cellular senescence and aging.Kaohsiung J Med Sci. 2010 Oct;26(10):515-31. doi: 10.1016/S1607-551X(10)70081-4. Kaohsiung J Med Sci. 2010. PMID: 20950777 Free PMC article. Review.
-
Transcriptomic analysis of maternally provisioned cues for phenotypic plasticity in the annual killifish, Austrofundulus limnaeus.Evodevo. 2017 Apr 21;8:6. doi: 10.1186/s13227-017-0069-7. eCollection 2017. Evodevo. 2017. PMID: 28439397 Free PMC article.
References
-
- Aasland R., Stewart, A.F., and Gibson, T. 1996. The SANT domain: A putative DNA-binding domain in the SWI-SNF and ADA complexes, the transcriptional co-repressor N-CoR and TFIIIB. Trends Biochem. Sci. 21: 87-88. - PubMed
-
- Akhmanova A.S., Bindels, P.C., Xu, J., Miedema, K., Kremer, H., and Hennig, W. 1995. Structure and expression of histone H3.3 genes in Drosophila melanogaster and Drosophila hydei. Genome 38: 586-600. - PubMed
-
- Ansieau S. and Leutz, A. 2002. The conserved Mynd domain of BS69 binds cellular and oncoviral proteins through a common PXLXP motif. J. Biol. Chem. 277: 4906-4910. - PubMed
-
- Beall E.L., Manak, J.R., Zhou, S., Bell, M., Lipsick, J.S., and Botchan, M.R. 2002. Role for a Drosophila Myb-containing protein complex in site-specific DNA replication. Nature 420: 833-837. - PubMed
-
- Boyer L.A., Langer, M.R., Crowley, K.A., Tan, S., Denu, J.M., and Peterson, C.L. 2002. Essential role for the SANT domain in the functioning of multiple chromatin remodeling enzymes. Mol. Cell 10: 935-942. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
