Delineation of xenobiotic substrate sites in rat glutathione S-transferase M1-1
- PMID: 16195544
- PMCID: PMC2253307
- DOI: 10.1110/ps.051651905
Delineation of xenobiotic substrate sites in rat glutathione S-transferase M1-1
Abstract
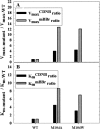
Glutathione S-transferases catalyze the conjugation of glutathione with endogenous and exogenous xenobiotics. Hu and Colman (1995) proposed that there are two distinct substrate sites in rat GST M1-1, a 1-chloro-2,4-dintrobenzene (CDNB) substrate site located in the vicinity of tyrosine-115, and a monobromobimane (mBBr) substrate site. To determine whether the mBBr substrate site is distinguishable from the CDNB substrate site, we tested S-(hydroxyethyl)bimane, a nonreactive derivative of mBBr, for its ability to compete kinetically with the substrates. We find that S-(hydroxyethyl)bimane is a competitive inhibitor (K(I) = 0.36 microM) when mBBr is used as substrate, but not when CDNB is used as substrate, demonstrating that these two sites are distinct. Using site-directed mutagenesis, we have localized the mBBr substrate site to an area midway through alpha-helix 4 (residues 90-114) and have identified residues that are important in the enzymatic reaction. Substitution of alanine at positions along alpha-helix 4 reveals that mutations at positions 103, 104, and 109 exhibit a greater perturbation of the enzymatic reaction with mBBr than with CDNB as substrate. Various other substitutions at positions 103 and 104 reveal that a hydrophobic residue is necessary at each of these positions to maintain optimal affinity of the enzyme for mBBr and preserve the secondary structure of the enzyme. Substitutions at position 109 indicate that this residue is important in the enzyme's affinity for mBBr but has a minimal effect on Vmax. These results demonstrate that the promiscuity of rat GST M1-1 is in part due to at least two distinct substrate sites.
Figures





Similar articles
-
Monobromobimane occupies a distinct xenobiotic substrate site in glutathione S-transferase pi.Protein Sci. 2003 Nov;12(11):2575-87. doi: 10.1110/ps.03249303. Protein Sci. 2003. PMID: 14573868 Free PMC article.
-
Probing the active site of alpha-class rat liver glutathione S-transferases using affinity labeling by monobromobimane.Protein Sci. 1997 Jan;6(1):43-52. doi: 10.1002/pro.5560060105. Protein Sci. 1997. PMID: 9007975 Free PMC article.
-
Monobromobimane as an affinity label of the xenobiotic binding site of rat glutathione S-transferase 3-3.J Biol Chem. 1995 Sep 15;270(37):21875-83. doi: 10.1074/jbc.270.37.21875. J Biol Chem. 1995. PMID: 7665611
-
The role of tyrosine-9 and the C-terminal helix in the catalytic mechanism of Alpha-class glutathione S-transferases.Biochem J. 1999 Nov 1;343 Pt 3(Pt 3):525-31. Biochem J. 1999. PMID: 10527929 Free PMC article.
-
Chemical arrows for enzymatic targets.FASEB J. 1997 Mar;11(4):217-26. doi: 10.1096/fasebj.11.4.9068610. FASEB J. 1997. PMID: 9068610 Review.
Cited by
-
Global liver gene expression differences in Nelore steers with divergent residual feed intake phenotypes.BMC Genomics. 2015 Mar 25;16(1):242. doi: 10.1186/s12864-015-1464-x. BMC Genomics. 2015. PMID: 25887532 Free PMC article.
-
Contribution of the mu loop to the structure and function of rat glutathione transferase M1-1.Protein Sci. 2006 Jun;15(6):1277-89. doi: 10.1110/ps.062129506. Epub 2006 May 2. Protein Sci. 2006. PMID: 16672236 Free PMC article.
-
Effect of Synthetic Peptides Identified in the Bullfrog Skin on Inflammation and Oxidative Stress Control: An In Vitro Analysis.Molecules. 2025 May 20;30(10):2223. doi: 10.3390/molecules30102223. Molecules. 2025. PMID: 40430395 Free PMC article.
-
The Redox System in C. elegans, a Phylogenetic Approach.J Toxicol. 2012;2012:546915. doi: 10.1155/2012/546915. Epub 2012 Jul 31. J Toxicol. 2012. PMID: 22899914 Free PMC article.
-
Ligandability Assessment of Human Glutathione Transferase M1-1 Using Pesticides as Chemical Probes.Int J Mol Sci. 2022 Mar 25;23(7):3606. doi: 10.3390/ijms23073606. Int J Mol Sci. 2022. PMID: 35408962 Free PMC article.
References
-
- Armstrong, R.N. 1991. Glutathione S-transferases: Reaction mechanism, structure, and function. Chem. Res. Toxicol. 4 131–140. - PubMed
-
- ———. 1998. Mechanistic imperative for the evolution of glutathione transferases. Curr. Opin. Chem. Biol. 2 618–623. - PubMed
-
- Barycki, J.J. and Colman, R.F. 1993. Affinity labeling of glutathione S-transferase, isozyme 4-4, by 4-(fluorosulfonyl)benzoic acid reveals Tyr 115 to be an important determinant of xenobiotic substrate specificity. Biochemistry 32 13002–13011. - PubMed
-
- Bhargava, M.M., Listowsky, I., and Arias, I.M. 1978. Studies on subunit structure and evidence that ligandin is a heterodimer. J. Biol. Chem. 253 4116–4119. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

