Evolutionary selection across the nuclear hormone receptor superfamily with a focus on the NR1I subfamily (vitamin D, pregnane X, and constitutive androstane receptors)
- PMID: 16197547
- PMCID: PMC1262763
- DOI: 10.1186/1478-1336-3-2
Evolutionary selection across the nuclear hormone receptor superfamily with a focus on the NR1I subfamily (vitamin D, pregnane X, and constitutive androstane receptors)
Abstract
Background: The nuclear hormone receptor (NR) superfamily complement in humans is composed of 48 genes with diverse roles in metabolic homeostasis, development, and detoxification. In general, NRs are strongly conserved between vertebrate species, and few examples of molecular adaptation (positive selection) within this superfamily have been demonstrated. Previous studies utilizing two-species comparisons reveal strong purifying (negative) selection of most NR genes, with two possible exceptions being the ligand-binding domains (LBDs) of the pregnane X receptor (PXR, NR1I2) and the constitutive androstane receptor (CAR, NR1I3), two proteins involved in the regulation of toxic compound metabolism and elimination. The aim of this study was to apply detailed phylogenetic analysis using maximum likelihood methods to the entire complement of genes in the vertebrate NR superfamily. Analyses were carried out both across all vertebrates and limited to mammals and also separately for the two major domains of NRs, the DNA-binding domain (DBD) and LBD, in addition to the full-length sequences. Additional functional data is also reported for activation of PXR and the vitamin D receptor (VDR; NR1I1) to gain further insight into the evolution of the NR1I subfamily.
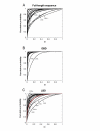
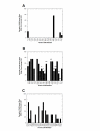
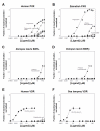
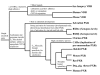
Results: The NR genes appear to be subject to strong purifying selection, particularly in the DBDs. Estimates of the ratio of the non-synonymous to synonymous nucleotide substitution rates (the omega ratio) revealed that only the PXR LBD had a sub-population of codons with an estimated omega ratio greater than 1. CAR was also unusual in showing high relative omega ratios in both the DBD and LBD, a finding that may relate to the recent appearance of the CAR gene (presumably by duplication of a pre-mammalian PXR gene) just prior to the evolution of mammals. Functional analyses of the NR1I subfamily show that human and zebrafish PXRs show similar activation by steroid hormones and early bile salts, properties not shared by sea lamprey, mouse, or human VDRs, or by Xenopus laevis PXRs.
Conclusion: NR genes generally show strong sequence conservation and little evidence for positive selection. The main exceptions are PXR and CAR, genes that may have adapted to cross-species differences in toxic compound exposure.
Figures




References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

