Identification of disease causing loci using an array-based genotyping approach on pooled DNA
- PMID: 16197552
- PMCID: PMC1262713
- DOI: 10.1186/1471-2164-6-138
Identification of disease causing loci using an array-based genotyping approach on pooled DNA
Abstract
Background: Pooling genomic DNA samples within clinical classes of disease followed by genotyping on whole-genome SNP microarrays, allows for rapid and inexpensive genome-wide association studies. Key to the success of these studies is the accuracy of the allelic frequency calculations, the ability to identify false-positives arising from assay variability and the ability to better resolve association signals through analysis of neighbouring SNPs.
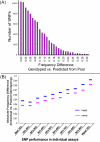
Results: We report the accuracy of allelic frequency measurements on pooled genomic DNA samples by comparing these measurements to the known allelic frequencies as determined by individual genotyping. We describe modifications to the calculation of k-correction factors from relative allele signal (RAS) values that remove biases and result in more accurate allelic frequency predictions. Our results show that the least accurate SNPs, those most likely to give false-positives in an association study, are identifiable by comparing their frequencies to both those from a known database of individual genotypes and those of the pooled replicates. In a disease with a previously identified genetic mutation, we demonstrate that one can identify the disease locus through the comparison of the predicted allelic frequencies in case and control pools. Furthermore, we demonstrate improved resolution of association signals using the mean of individual test-statistics for consecutive SNPs windowed across the genome. A database of k-correction factors for predicting allelic frequencies for each SNP, derived from several thousand individually genotyped samples, is provided. Lastly, a Perl script for calculating RAS values for the Affymetrix platform is provided.
Conclusion: Our results illustrate that pooling of DNA samples is an effective initial strategy to identify a genetic locus. However, it is important to eliminate inaccurate SNPs prior to analysis by comparing them to a database of individually genotyped samples as well as by comparing them to replicates of the pool. Lastly, detection of association signals can be improved by incorporating data from neighbouring SNPs.
Figures


References
-
- Matsuzaki H, Dong S, Loi H, Di X, Liu G, Hubbell E, Law J, Berntsen T, Chadha M, Hui H, Yang G, Kennedy GC, Webster TA, Cawley S, Walsh E, Jones KW, Fodor SP, Mei R. Genotyping over 100,000 SNPs on a pair of olignucleotide arrays. Nature Methods. 2004;1:109–111. doi: 10.1038/nmeth718. - DOI - PubMed
-
- Fan JB, Oliphant A, Shen R, Kermani BG, Garcia F, Gunderson KL, Hansen M, Steemers F, Butler SL, Deloukas P, Galver L, Hunt S, McBride C, Bibikova M, Rubano T, Chen J, Wickham E, Doucet D, Chang W, Campbell D, Zhang B, Kruglyak S, Bentley D, Haas J, Rigault P, Zhou L, Stuelpnagel J, Chee MS. Highly parallel SNP genotyping. Cold Spring Harb Symp Quant Biol. 2003;68:69–78. doi: 10.1101/sqb.2003.68.69. - DOI - PubMed
-
- Marnellos G. High-throughput SNP analysis for genetic association studies. Curr Opin Drug Discov Devel. 2003;6:317–321. - PubMed
-
- Risch N, Teng J. The relative power of family-based and case-control designs for linkage disequilibrium studies of complex human diseases I. DNA pooling. Genome Res. 1998;8:1273–1288. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

