Fluorescence correlation spectroscopy diffusion laws to probe the submicron cell membrane organization
- PMID: 16199500
- PMCID: PMC1366968
- DOI: 10.1529/biophysj.105.067959
Fluorescence correlation spectroscopy diffusion laws to probe the submicron cell membrane organization
Abstract
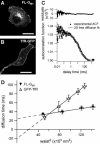
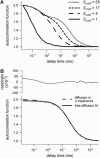
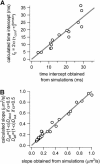
To probe the complexity of the cell membrane organization and dynamics, it is important to obtain simple physical observables from experiments on live cells. Here we show that fluorescence correlation spectroscopy (FCS) measurements at different spatial scales enable distinguishing between different submicron confinement models. By plotting the diffusion time versus the transverse area of the confocal volume, we introduce the so-called FCS diffusion law, which is the key concept throughout this article. First, we report experimental FCS diffusion laws for two membrane constituents, which are respectively a putative raft marker and a cytoskeleton-hindered transmembrane protein. We find that these two constituents exhibit very distinct behaviors. To understand these results, we propose different models, which account for the diffusion of molecules either in a membrane comprising isolated microdomains or in a meshwork. By simulating FCS experiments for these two types of organization, we obtain FCS diffusion laws in agreement with our experimental observations. We also demonstrate that simple observables derived from these FCS diffusion laws are strongly related to confinement parameters such as the partition of molecules in microdomains and the average confinement time of molecules in a microdomain or a single mesh of a meshwork.
Figures









Comment in
-
Fluorescence corralation spectroscopy.Biophys J. 2005 Dec;89(6):3678-9. doi: 10.1529/biophysj.105.074161. Epub 2005 Sep 30. Biophys J. 2005. PMID: 16199507 Free PMC article. No abstract available.
References
-
- Jacobson, K., E. D. Sheets, and R. Simson. 1995. Revisiting the fluid mosaic model of membranes. Science. 268:1441–1442. - PubMed
-
- Singer, S. J., and G. L. Nicolson. 1972. The fluid mosaic model of the structure of cell membranes. Science. 175:720–731. - PubMed
-
- Brown, D. A., and J. K. Rose. 1992. Sorting of GPI-anchored proteins to glycolipid-enriched membrane subdomains during transport to the apical cell surface. Cell. 68:533–544. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

