Prenatal detection of unbalanced chromosomal rearrangements by array CGH
- PMID: 16199537
- PMCID: PMC2563226
- DOI: 10.1136/jmg.2005.037648
Prenatal detection of unbalanced chromosomal rearrangements by array CGH
Abstract
Background: Karyotype analysis has been the standard method for prenatal cytogenetic diagnosis since the 1970s. Although highly reliable, the major limitation remains the requirement for cell culture, resulting in a delay of as much as 14 days to obtaining test results. Fluorescent in situ hybridisation (FISH) and quantitative fluorescent PCR (QF-PCR) rapidly detect common chromosomal abnormalities but do not provide a genome wide screen for unexpected imbalances. Array comparative genomic hybridisation (CGH) has the potential to combine the speed of DNA analysis with a large capacity to scan for genomic abnormalities. We have developed a genomic microarray of approximately 600 large insert clones designed to detect aneuploidy, known microdeletion syndromes, and large unbalanced chromosomal rearrangements.
Methods: This array was tested alongside an array with an approximate resolution of 1 Mb in a blind study of 30 cultured prenatal and postnatal samples with microscopically confirmed unbalanced rearrangements.
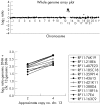
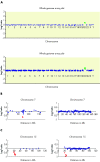
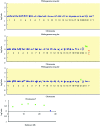
Results: At 1 Mb resolution, 22/30 rearrangements were identified, whereas 29/30 aberrations were detected using the custom designed array, owing to the inclusion of specifically chosen clones to give increased resolution at genomic loci clinically implicated in known microdeletion syndromes. Both arrays failed to identify a triploid karyotype. Thirty normal control samples produced no false positive results.
Conclusions: Analysis of 30 uncultured prenatal samples showed that array CGH is capable of detecting aneuploidy in DNA isolated from as little as 1 ml of uncultured amniotic fluid; 29/30 samples were correctly diagnosed, the exception being another case of triploidy. These studies demonstrate the potential for array CGH to replace conventional cytogenetics in the great majority of prenatal diagnosis cases.
Conflict of interest statement
Competing interests: there are no competing interests.
References
-
- Waters J J, Waters K S. Trends in cytogenetic prenatal diagnosis in the UK: results from UKNEQAS external audit, 1987–1998. Prenat Diagn 1999191023–1026. - PubMed
-
- Caspersson T, Farber S, Foley G E, Kudynowski J, Modest E J, Simonsson E, Wagh U, Zech L. Chemical differentiation along metaphase chromosomes. Exp Cell Res 196849219–222. - PubMed
-
- Shaffer L G, Bejjani B A. A cytogeneticist's perspective on genomic microarrays. Hum Reprod Update 200410221–226. - PubMed
-
- Shaffer L G, Lupski J R. Molecular mechanisms for constitutional chromosomal rearrangements in humans. Annu Rev Genet 200034297–329. - PubMed
-
- Cirigliano V, Voglino G, Canadas M P, Marongiu A, Ejarque M, Ordonez E, Plaja A, Massobrio M, Todros T, Fuster C, Campogrande M, Egozcue J, Adinolfi M. Rapid prenatal diagnosis of common chromosome aneuploidies by QF‐PCR. Assessment on 18,000 consecutive clinical samples. Mol Hum Reprod 200410839–846. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous