The lrp gene and its role in type I fimbriation in Citrobacter rodentium
- PMID: 16199571
- PMCID: PMC1251604
- DOI: 10.1128/JB.187.20.7009-7017.2005
The lrp gene and its role in type I fimbriation in Citrobacter rodentium
Abstract
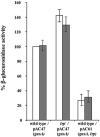
Citrobacter rodentium is a murine pathogen that is now widely used as an in vivo model for gastrointestinal infections due to its similarities with human enteropathogens, such as the possession of a locus for enterocyte effacement (the LEE island). We studied the lrp gene of C. rodentium and found that it encodes a product highly similar to members of the Lrp (leucine-responsive regulatory protein) family of transcriptional regulators, able to recognize leucine as an effector and to repress the expression of its own structural gene. In enterobacteria, Lrp is a global regulator of gene expression, as it controls a large variety of genes, including those coding for cell appendages and other potential virulence factors. Based on the well-established role of Lrp on the expression of pilus genes in Escherichia coli, we also studied the role of Lrp in controlling the formation of the type I pilus in C. rodentium. Type I pili, produced by the fim system, are virulence factors of uropathogens, involved in mediating bacterial adhesion to bladder epithelial cells. Yeast agglutination assays showed that Lrp is needed for type I pilus formation and real-time PCR experiments indicated that Lrp has a strong leucine-mediated effect on the expression of the fimAICDFGH operon. Mutant studies indicated that this positive action is exerted mainly through a positive control of Lrp on the phase variation mechanism that regulates fimAICDFGH expression. A quantitative analysis of its expression suggested that this operon may also be negatively regulated at the level of transcription.
Figures

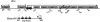


Similar articles
-
Direct and indirect control of Lrp on LEE pathogenicity genes of Citrobacter rodentium.FEMS Microbiol Lett. 2011 Dec;325(1):64-70. doi: 10.1111/j.1574-6968.2011.02411.x. Epub 2011 Oct 3. FEMS Microbiol Lett. 2011. PMID: 22092863
-
A positive regulatory loop controls expression of the locus of enterocyte effacement-encoded regulators Ler and GrlA.J Bacteriol. 2005 Dec;187(23):7918-30. doi: 10.1128/JB.187.23.7918-7930.2005. J Bacteriol. 2005. PMID: 16291665 Free PMC article.
-
A nucleoprotein activation complex between the leucine-responsive regulatory protein and DNA upstream of the gltBDF operon in Escherichia coli.J Mol Biol. 1997 Jul 11;270(2):152-68. doi: 10.1006/jmbi.1997.1057. J Mol Biol. 1997. PMID: 9236118
-
Leucine-responsive regulatory protein: a global regulator of gene expression in E. coli.Annu Rev Microbiol. 1995;49:747-75. doi: 10.1146/annurev.mi.49.100195.003531. Annu Rev Microbiol. 1995. PMID: 8561478 Review.
-
The intricate workings of a bacterial epigenetic switch.Adv Exp Med Biol. 2004;547:83-9. doi: 10.1007/978-1-4419-8861-4_7. Adv Exp Med Biol. 2004. PMID: 15230094 Review.
Cited by
-
The leucine-responsive regulatory proteins/feast-famine regulatory proteins: an ancient and complex class of transcriptional regulators in bacteria and archaea.Crit Rev Biochem Mol Biol. 2021 Aug;56(4):373-400. doi: 10.1080/10409238.2021.1925215. Epub 2021 Jun 20. Crit Rev Biochem Mol Biol. 2021. PMID: 34151666 Free PMC article. Review.
-
Inactivation of MSMEG_0412 gene drastically affects surface related properties of Mycobacterium smegmatis.BMC Microbiol. 2016 Nov 8;16(1):267. doi: 10.1186/s12866-016-0888-z. BMC Microbiol. 2016. PMID: 27825305 Free PMC article.
-
Surface Bacterioplankton Community Structure Crossing the Antarctic Circumpolar Current Fronts.Microorganisms. 2023 Mar 9;11(3):702. doi: 10.3390/microorganisms11030702. Microorganisms. 2023. PMID: 36985275 Free PMC article.
-
Transcriptional analysis of the recA gene of Streptococcus thermophilus.Microb Cell Fact. 2006 Sep 14;5:29. doi: 10.1186/1475-2859-5-29. Microb Cell Fact. 2006. PMID: 16972988 Free PMC article.
-
Mapping the microbial diversity associated with different geochemical regimes in the shallow-water hydrothermal vents of the Aeolian archipelago, Italy.Front Microbiol. 2023 Aug 11;14:1134114. doi: 10.3389/fmicb.2023.1134114. eCollection 2023. Front Microbiol. 2023. PMID: 37637107 Free PMC article.
References
-
- Brinkman, A. B., T. J. Ettema, W. M. de Vos, and J. van der Oost. 2003. The Lrp family of transcriptional regulators. Mol. Microbiol. 48:287-294. - PubMed
-
- Camacho, E. M., and J. Casadesus. 2002. Conjugal transfer of the virulence plasmid of Salmonella enterica is regulated by the leucine-responsive regulatory protein and DNA adenine methylation. Mol. Microbiol. 44:1589-1598. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

