Transcriptional program of early sporulation and stationary-phase events in Clostridium acetobutylicum
- PMID: 16199581
- PMCID: PMC1251621
- DOI: 10.1128/JB.187.20.7103-7118.2005
Transcriptional program of early sporulation and stationary-phase events in Clostridium acetobutylicum
Abstract
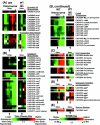
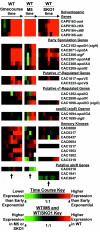
DNA microarray analysis of Clostridium acetobutylicum was used to examine the genomic-scale gene expression changes during the shift from exponential-phase growth and acidogenesis to stationary phase and solventogenesis. Self-organizing maps were used to identify novel expression patterns of functional gene classes, including aromatic and branched-chain amino acid synthesis, ribosomal proteins, cobalt and iron transporters, cobalamin biosynthesis, and lipid biosynthesis. The majority of pSOL1 megaplasmid genes (in addition to the solventogenic genes aad-ctfA-ctfB and adc) had increased expression at the onset of solventogenesis, suggesting that other megaplasmid genes may play a role in stationary-phase phenomena. Analysis of sporulation genes and comparison with published Bacillus subtilis results indicated conserved expression patterns of early sporulation genes, including spo0A, the sigF operon, and putative canonical genes of the sigma(H) and sigma(F) regulons. However, sigE expression could not be detected within 7.5 h of initial spo0A expression, consistent with the observed extended time between the appearance of clostridial forms and endospore formation. The results were compared with microarray comparisons of the wild-type strain and the nonsolventogenic, asporogenous M5 strain, which lacks the pSOL1 megaplasmid. While some results were similar, the expression of primary metabolism genes and heat shock proteins was higher in M5, suggesting a difference in metabolic regulation or a butyrate stress response in M5. The results of this microarray platform and analysis were further validated by comparing gene expression patterns to previously published Northern analyses, reporter assays, and two-dimensional protein electrophoresis data of metabolic genes (including all major solventogenesis genes), sporulation genes, heat shock proteins, and other solventogenesis-induced gene expression.
Figures
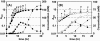



References
-
- Alsaker, K. V., C. J. Paredes, and E. T. Papoutsakis. Design, optimization and validation of genomic DNA microarrays for examining the Clostridium acetobutylicum transcriptome. Biotechnol. Bioprocess Eng., in press.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

