Global transcriptome analysis of Shewanella oneidensis MR-1 exposed to different terminal electron acceptors
- PMID: 16199584
- PMCID: PMC1251602
- DOI: 10.1128/JB.187.20.7138-7145.2005
Global transcriptome analysis of Shewanella oneidensis MR-1 exposed to different terminal electron acceptors
Abstract
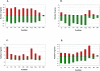
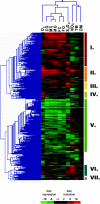
To gain insight into the complex structure of the energy-generating networks in the dissimilatory metal reducer Shewanella oneidensis MR-1, global mRNA patterns were examined in cells exposed to a wide range of metal and non-metal electron acceptors. Gene expression patterns were similar irrespective of which metal ion was used as electron acceptor, with 60% of the differentially expressed genes showing similar induction or repression relative to fumarate-respiring conditions. Several groups of genes exhibited elevated expression levels in the presence of metals, including those encoding putative multidrug efflux transporters, detoxification proteins, extracytoplasmic sigma factors and PAS-domain regulators. Only one of the 42 predicted c-type cytochromes in MR-1, SO3300, displayed significantly elevated transcript levels across all metal-reducing conditions. Genes encoding decaheme cytochromes MtrC and MtrA that were previously linked to the reduction of different forms of Fe(III) and Mn(IV), exhibited only slight decreases in relative mRNA abundances under metal-reducing conditions. In contrast, specific transcriptome responses were displayed to individual non-metal electron acceptors resulting in the identification of unique groups of nitrate-, thiosulfate- and TMAO-induced genes including previously uncharacterized multi-cytochrome gene clusters. Collectively, the gene expression results reflect the fundamental differences between metal and non-metal respiratory pathways of S. oneidensis MR-1, where the coordinate induction of detoxification and stress response genes play a key role in adaptation of this organism under metal-reducing conditions. Moreover, the relative paucity and/or the constitutive nature of genes involved in electron transfer to metals is likely due to the low-specificity and the opportunistic nature of the metal-reducing electron transport pathways.
Figures

References
-
- Anderson, R. T., H. A. Vrionis, I. Ortiz-Bernad, C. T. Resch, P. E. Long, R. Dayvault, K. Karp, S. Marutzky, D. R. Metzler, A. Peacock, D. C. White, M. Lowe, and D. R. Lovley. 2003. Stimulating the in situ activity of Geobacter species to remove uranium from the groundwater of a uranium-contaminated aquifer. Appl. Environ. Microbiol. 69:5884-5891. - PMC - PubMed
-
- Beliaev, A. S., D. A. Saffarini, J. L. McLaughlin, and D. Hunnicutt. 2001. MtrC, an outer membrane decahaem c cytochrome required for metal reduction in Shewanella putrefaciens MR-1. Mol. Microbiol. 39:722-730. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

