Dcp2 Decaps m2,2,7GpppN-capped RNAs, and its activity is sequence and context dependent
- PMID: 16199859
- PMCID: PMC1265788
- DOI: 10.1128/MCB.25.20.8779-8791.2005
Dcp2 Decaps m2,2,7GpppN-capped RNAs, and its activity is sequence and context dependent
Abstract
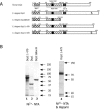
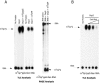
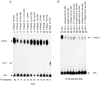
Hydrolysis of the mRNA cap plays a pivotal role in initiating and completing mRNA turnover. In nematodes, mRNA metabolism and cap-interacting proteins must deal with two populations of mRNAs, spliced leader trans-spliced mRNAs with a trimethylguanosine cap and non-trans-spliced mRNAs with a monomethylguanosine cap. We describe here the characterization of nematode Dcp1 and Dcp2 proteins. Dcp1 was inactive in vitro on both free cap and capped RNA and did not significantly enhance Dcp2 activity. Nematode Dcp2 is an RNA-decapping protein that does not bind cap and is not inhibited by cap analogs but is effectively inhibited by competing RNA irrespective of RNA sequence and cap. Nematode Dcp2 activity is influenced by both 5' end sequence and its context. The trans-spliced leader sequence on mRNAs reduces Dcp2 activity approximately 10-fold, suggesting that 5'-to-3' turnover of trans-spliced RNAs may be regulated. Nematode Dcp2 decaps both m(7)GpppG- and m(2,2,7)GpppG-capped RNAs. Surprisingly, both budding yeast and human Dcp2 are also active on m(2,2,7)GpppG-capped RNAs. Overall, the data suggest that Dcp2 activity can be influenced by both sequence and context and that Dcp2 may contribute to gene regulation in multiple RNA pathways, including monomethyl- and trimethylguanosine-capped RNAs.
Figures




References
-
- Baker, K. E., and R. Parker. 2004. Nonsense-mediated mRNA decay: terminating erroneous gene expression. Curr. Opin. Cell Biol. 16:293-299. - PubMed
-
- Beelman, C. A., A. Stevens, G. Caponigro, T. E. LaGrandeur, L. Hatfield, D. M. Fortner, and R. Parker. 1996. An essential component of the decapping enzyme required for normal rates of mRNA turnover. Nature 382:642-646. - PubMed
-
- Blumenthal, T. 1995. Trans-splicing and polycistronic transcription in Caenorhabditis elegans. Trends Genet. 11:132-136. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
