Nucleophosmin is required for DNA integrity and p19Arf protein stability
- PMID: 16199867
- PMCID: PMC1265791
- DOI: 10.1128/MCB.25.20.8874-8886.2005
Nucleophosmin is required for DNA integrity and p19Arf protein stability
Abstract
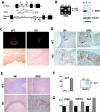
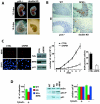
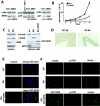
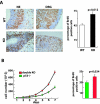
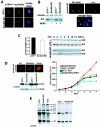
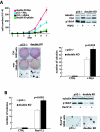
Nucleophosmin (NPM) is a nucleolar phosphoprotein that binds the tumor suppressors p53 and p19(Arf) and is thought to be indispensable for ribogenesis, cell proliferation, and survival after DNA damage. The NPM gene is the most frequent target of genetic alterations in leukemias and lymphomas, though its role in tumorigenesis is unknown. We report here the first characterization of a mouse NPM knockout strain. Lack of NPM expression results in accumulation of DNA damage, activation of p53, widespread apoptosis, and mid-stage embryonic lethality. Fibroblasts explanted from null embryos fail to grow and rapidly acquire a senescent phenotype. Transfer of the NPM mutation into a p53-null background rescued apoptosis in vivo and fibroblast proliferation in vitro. Cells null for both p53 and NPM grow faster than control cells and are more susceptible to transformation by activated oncogenes, such as mutated Ras or overexpressed Myc. In the absence of NPM, Arf protein is excluded from nucleoli and is markedly less stable. Our data demonstrate that NPM regulates DNA integrity and, through Arf, inhibits cell proliferation and are consistent with a putative tumor-suppressive function of NPM.
Figures
References
-
- Bartek, J., C. lukas, and J. Lukas. 2004. Checking on DNA damage in S phase. Nat. Rev. Mol. Cell. Biol. 5:792-804. - PubMed
-
- Bartkova, J., Z. Horejsi, K. Koed, A. Kramer, F. Tort, K. Zieger, P. Guldberg, M. Sehested, J. M. Nesland, C. Lukas, T. Orntoft, J. Lukas, and J. Bartek. 2005. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature 434:864-870. - PubMed
-
- Borer, R. A., C. F. Lehner, H. M. Eppenberger, and E. A. Nigg. 1989. Major nucleolar proteins shuttle between nucleus and cytoplasm. Cell 56:379-390. - PubMed
-
- Borggrefe, T., M. Wabl, A. T. Akhmedov, and R. Jessberger. 1998. A B-cell-specific DNA recombination complex. J. Biol. Chem. 273:17025-17035. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
