Environments shape the nucleotide composition of genomes
- PMID: 16200051
- PMCID: PMC1369203
- DOI: 10.1038/sj.embor.7400538
Environments shape the nucleotide composition of genomes
Abstract
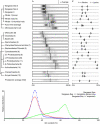
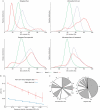
To test the impact of environments on genome evolution, we analysed the relative abundance of the nucleotides guanine and cytosine ('GC content') of large numbers of sequences from four distinct environmental samples (ocean surface water, farm soil, an acidophilic mine drainage biofilm and deep-sea whale carcasses). We show that the GC content of complex microbial communities seems to be globally and actively influenced by the environment. The observed nucleotide compositions cannot be easily explained by distinct phylogenetic origins of the species in the environments; the genomic GC content may change faster than was previously thought, and is also reflected in the amino-acid composition of the proteins in these habitats.
Figures
References
-
- Bentley SD, Parkhill J (2004) Comparative genomic structure of prokaryotes. Annu Rev Genet 38: 771–792 - PubMed
-
- Bharanidharan D, Bhargavi GR, Uthanumallian K, Gautham N (2004) Correlations between nucleotide frequencies and amino acid composition in 115 bacterial species. Biochem Biophys Res Commun 315: 1097–1103 - PubMed
-
- Delong EF (2005) Microbial community genomics in the ocean. Nat Rev Microbiol 6: 459–469 - PubMed
-
- Fickett JW (1995) ORFs and genes: how strong a connection? J Comput Biol 2: 117–123 - PubMed
-
- Foster PG, Jermiin LS, Hickey DA (1997) Nucleotide composition bias affects amino acid content in proteins coded by animal mitochondria. J Mol Evol 44: 282–288 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

