The Amber biomolecular simulation programs
- PMID: 16200636
- PMCID: PMC1989667
- DOI: 10.1002/jcc.20290
The Amber biomolecular simulation programs
Abstract
We describe the development, current features, and some directions for future development of the Amber package of computer programs. This package evolved from a program that was constructed in the late 1970s to do Assisted Model Building with Energy Refinement, and now contains a group of programs embodying a number of powerful tools of modern computational chemistry, focused on molecular dynamics and free energy calculations of proteins, nucleic acids, and carbohydrates.
(c) 2005 Wiley Periodicals, Inc.
Figures



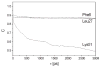
References
-
- Weiner P, Kollman P. J Comput Chem. 1981;2:287.
-
- Pearlman D, Case D, Caldwell J, Ross W, Cheatham T, III, DeBolt S, Ferguson D, Seibel G, Kollman P. Comp Phys Commun. 1995;91:1.
-
- Allen, M.; Tildesley, D. Computer Simulation of Liquids; Clarendon Press: Oxford, 1987.
-
- Frenkel, D.; Smit, B. Understanding Molecular Simulation: From Algorithms to Applications; Academic Press: San Diego, 1996.
-
- Becker, O.; MacKerell, A.; Roux, B.; Watanabe, M. Computational Biochemistry and Biophysics; Marcel Dekker: New York, 2001.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

