Emergence of young human genes after a burst of retroposition in primates
- PMID: 16201836
- PMCID: PMC1251493
- DOI: 10.1371/journal.pbio.0030357
Emergence of young human genes after a burst of retroposition in primates
Abstract
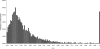
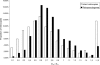
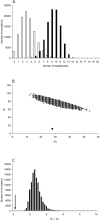
The origin of new genes through gene duplication is fundamental to the evolution of lineage- or species-specific phenotypic traits. In this report, we estimate the number of functional retrogenes on the lineage leading to humans generated by the high rate of retroposition (retroduplication) in primates. Extensive comparative sequencing and expression studies coupled with evolutionary analyses and simulations suggest that a significant proportion of recent retrocopies represent bona fide human genes. We estimate that at least one new retrogene per million years emerged on the human lineage during the past approximately 63 million years of primate evolution. Detailed analysis of a subset of the data shows that the majority of retrogenes are specifically expressed in testis, whereas their parental genes show broad expression patterns. Consistently, most retrogenes evolved functional roles in spermatogenesis. Proteins encoded by X chromosome-derived retrogenes were strongly preserved by purifying selection following the duplication event, supporting the view that they may act as functional autosomal substitutes during X-inactivation of late spermatogenesis genes. Also, some retrogenes acquired a new or more adapted function driven by positive selection. We conclude that retroduplication significantly contributed to the formation of recent human genes and that most new retrogenes were progressively recruited during primate evolution by natural and/or sexual selection to enhance male germline function.
Figures

References
-
- Samonte RV, Eichler EE. Segmental duplications and the evolution of the primate genome. Nat Rev Genet. 2002;3:65–72. - PubMed
-
- Brosius J. Retroposons—Seeds of evolution. Science. 1991;251:753. - PubMed
-
- Long M, Betran E, Thornton K, Wang W. The origin of new genes: Glimpses from the young and old. Nat Rev Genet. 2003;4:865–875. - PubMed
-
- Mighell AJ, Smith NR, Robinson PA, Markham AF. Vertebrate pseudogenes. FEBS Lett. 2000;468:109–114. - PubMed
-
- Emerson JJ, Kaessmann H, Betran E, Long M. Extensive gene traffic on the mammalian X chromosome. Science. 2004;303:537–540. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

