Physical and functional interactions between Escherichia coli MutY and endonuclease VIII
- PMID: 16201966
- PMCID: PMC1383697
- DOI: 10.1042/BJ20051133
Physical and functional interactions between Escherichia coli MutY and endonuclease VIII
Abstract
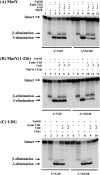
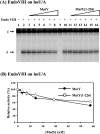
Both GO (7,8-dihydro-8-oxoguanine) and hoU (5-hydroxyuracil) are highly mutagenic because DNA polymerase frequently misincorporates adenine opposite these damaged bases. In Escherichia coli, MutY DNA glycosylase can remove misincorporated adenine opposite G or GO on the template strand during DNA replication. MutY remains bound to the product that contains an AP (apurinic/apyrimidinic) site. Endo VIII (endonuclease VIII) can remove oxidized pyrimidine and weakly remove GO by its DNA glycosylase and beta/delta-elimination activities. In the present paper, we demonstrate that Endo VIII can promote MutY dissociation from AP/G, but not from AP/GO, and can promote beta/delta-elimination on the products of MutY. MutY interacts physically with Endo VIII through its C-terminal domain. MutY has a moderate affinity for DNA containing a hoU/A mismatch, which is a substrate of Endo VIII. MutY competes with Endo VIII and inhibits Endo VIII activity on DNA that contains a hoU/A mismatch. Moreover, MutY has a weak adenine glycosylase activity on hoU/A mismatches. These results suggest that MutY may have some role in reducing the mutagenic effects of hoU.
Figures

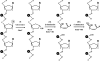


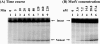
Similar articles
-
Escherichia coli apurinic-apyrimidinic endonucleases enhance the turnover of the adenine glycosylase MutY with G:A substrates.J Biol Chem. 2002 Jun 21;277(25):22605-15. doi: 10.1074/jbc.M203037200. Epub 2002 Apr 17. J Biol Chem. 2002. PMID: 11960995
-
Multiple DNA glycosylases for repair of 8-oxoguanine and their potential in vivo functions.Prog Nucleic Acid Res Mol Biol. 2001;68:193-205. doi: 10.1016/s0079-6603(01)68100-5. Prog Nucleic Acid Res Mol Biol. 2001. PMID: 11554297 Review.
-
Differential specificity of human and Escherichia coli endonuclease III and VIII homologues for oxidative base lesions.J Biol Chem. 2004 Apr 2;279(14):14464-71. doi: 10.1074/jbc.M400393200. Epub 2004 Jan 20. J Biol Chem. 2004. PMID: 14734554
-
A novel role for Escherichia coli endonuclease VIII in prevention of spontaneous G-->T transversions.J Bacteriol. 1999 Oct;181(20):6396-402. doi: 10.1128/JB.181.20.6396-6402.1999. J Bacteriol. 1999. PMID: 10515930 Free PMC article.
-
Potential double-flipping mechanism by E. coli MutY.Prog Nucleic Acid Res Mol Biol. 2001;68:349-64. doi: 10.1016/s0079-6603(01)68111-x. Prog Nucleic Acid Res Mol Biol. 2001. PMID: 11554310 Review.
Cited by
-
MUTYH DNA glycosylase: the rationale for removing undamaged bases from the DNA.Front Genet. 2013 Feb 28;4:18. doi: 10.3389/fgene.2013.00018. eCollection 2013. Front Genet. 2013. PMID: 23450852 Free PMC article.
-
Physical and functional interactions between Escherichia coli MutY glycosylase and mismatch repair protein MutS.J Bacteriol. 2007 Feb;189(3):902-10. doi: 10.1128/JB.01513-06. Epub 2006 Nov 17. J Bacteriol. 2007. PMID: 17114250 Free PMC article.
-
Analysis of the impact of a uracil DNA glycosylase attenuated in AP-DNA binding in maintenance of the genomic integrity in Escherichia coli.Nucleic Acids Res. 2010 Apr;38(7):2291-301. doi: 10.1093/nar/gkp1210. Epub 2010 Jan 7. Nucleic Acids Res. 2010. PMID: 20056657 Free PMC article.
-
Metagenome mining and functional analysis reveal oxidized guanine DNA repair at the Lost City Hydrothermal Field.PLoS One. 2024 May 8;19(5):e0284642. doi: 10.1371/journal.pone.0284642. eCollection 2024. PLoS One. 2024. PMID: 38718041 Free PMC article.
References
-
- Halliwell B., Gutteridge J. M. New York: Oxford University Press; 1989. Free Radicals in Biology and Medicine.
-
- Tchou J., Grollman A. P. Repair of DNA containing the oxidatively-damaged base 8-hydroxyguanine. Mutat. Res. 1993;299:277–287. - PubMed
-
- Lu A.-L., Li X., Gu Y., Wright P. M., Chang D.-Y. Repair of oxidative DNA damage. Cell Biochem. Biophys. 2001;35:141–170. - PubMed
-
- Maki H., Sekiguchi M. MutT protein specifically hydrolyses a potent mutagenic substrate for DNA synthesis. Nature (London) 1992;355:273–275. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

