Identification of programmed translational -1 frameshifting sites in the genome of Saccharomyces cerevisiae
- PMID: 16204194
- PMCID: PMC1240084
- DOI: 10.1101/gr.4258005
Identification of programmed translational -1 frameshifting sites in the genome of Saccharomyces cerevisiae
Retraction in
-
Identification of programmed translational -1 frameshifting sites in the genome of Saccharomyces cerevisiae.Genome Res. 2006 Aug;16(8):1074. Genome Res. 2006. PMID: 16882864 Free PMC article. No abstract available.
Abstract
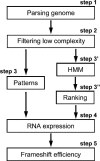
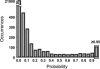
Frameshifting is a recoding event that allows the expression of two polypeptides from the same mRNA molecule. Most recoding events described so far are used by viruses and transposons to express their replicase protein. The very few number of cellular proteins known to be expressed by a -1 ribosomal frameshifting has been identified by chance. The goal of the present work was to set up a systematic strategy, based on complementary bioinformatics, molecular biology, and functional approaches, without a priori knowledge of the mechanism involved. Two independent methods were devised. The first looks for genomic regions in which two ORFs, each carrying a protein pattern, are in a frameshifted arrangement. The second uses Hidden Markov Models and likelihood in a two-step approach. When this strategy was applied to the Saccharomyces cerevisiae genome, 189 candidate regions were found, of which 58 were further functionally investigated. Twenty-eight of them expressed a full-length mRNA covering the two ORFs, and 11 showed a -1 frameshift efficiency varying from 5% to 13% (50-fold higher than background), some of which corresponds to genes with known functions. From other ascomycetes, four frameshifted ORFs are found fully conserved. Strikingly, most of the candidates do not display a classical viral-like frameshift signal and would have escaped a search based on current models of frameshifting. These results strongly suggest that -1 frameshifting might be more widely distributed than previously thought.
Figures




Similar articles
-
Characterization of new proteins found by analysis of short open reading frames from the full yeast genome.Yeast. 1997 Nov;13(14):1363-74. doi: 10.1002/(SICI)1097-0061(199711)13:14<1363::AID-YEA182>3.0.CO;2-8. Yeast. 1997. PMID: 9392081
-
Identification of putative regulatory upstream ORFs in the yeast genome using heuristics and evolutionary conservation.BMC Bioinformatics. 2007 Aug 8;8:295. doi: 10.1186/1471-2105-8-295. BMC Bioinformatics. 2007. PMID: 17686169 Free PMC article.
-
How many protein-coding genes are there in the Saccharomyces cerevisiae genome?Yeast. 2002 May;19(7):619-29. doi: 10.1002/yea.865. Yeast. 2002. PMID: 11967832
-
[Decade of genomics--methods for genome investigation in yeast Saccharomyces cerevisiae].Postepy Biochem. 2006;52(4):435-47. Postepy Biochem. 2006. PMID: 17536513 Review. Polish.
-
Programmed translational frameshifting.Annu Rev Genet. 1996;30:507-28. doi: 10.1146/annurev.genet.30.1.507. Annu Rev Genet. 1996. PMID: 8982463 Review.
Cited by
-
Specific effects of ribosome-tethered molecular chaperones on programmed -1 ribosomal frameshifting.Eukaryot Cell. 2006 Apr;5(4):762-70. doi: 10.1128/EC.5.4.762-770.2006. Eukaryot Cell. 2006. PMID: 16607023 Free PMC article.
-
High Diversity of Novel Viruses in the Tree Pathogen Phytophthora castaneae Revealed by High-Throughput Sequencing of Total and Small RNA.Front Microbiol. 2022 Jun 16;13:911474. doi: 10.3389/fmicb.2022.911474. eCollection 2022. Front Microbiol. 2022. PMID: 35783401 Free PMC article.
-
Multiple Viral Infections Detected in Phytophthora condilina by Total and Small RNA Sequencing.Viruses. 2021 Apr 4;13(4):620. doi: 10.3390/v13040620. Viruses. 2021. PMID: 33916635 Free PMC article.
-
Control of gene expression by translational recoding.Adv Protein Chem Struct Biol. 2012;86:129-49. doi: 10.1016/B978-0-12-386497-0.00004-9. Adv Protein Chem Struct Biol. 2012. PMID: 22243583 Free PMC article. Review.
-
Core gene set of the species Saccharomyces cerevisiae.bioRxiv [Preprint]. 2025 Apr 30:2023.09.07.545205. doi: 10.1101/2023.09.07.545205. bioRxiv. 2025. PMID: 40502033 Free PMC article. Preprint.
References
WEB SITE REFERENCES
-
- http://bioperl.org/; The Bioperl Project.
-
- http://cbi.labri.fr/Genolevures/; Genolevure.
-
- http://www.lri.fr/~denise/GenRGenS/; GenRGenS home page.
-
- http://www.ebi.ac.uk/interpro/; InterPro database.
-
- http://mips.gsf.de/genre/proj/yeast/; Munish information center for protein sequences (MIPS).
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
