Chromosome inversions, local adaptation and speciation
- PMID: 16204214
- PMCID: PMC1461441
- DOI: 10.1534/genetics.105.047985
Chromosome inversions, local adaptation and speciation
Erratum in
-
Chromosome Inversions, Local Adaptation and Speciation.Genetics. 2018 Jan;208(1):433. doi: 10.1534/genetics.117.300572. Genetics. 2018. PMID: 29301950 Free PMC article. No abstract available.
Abstract
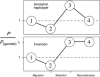
We study the evolution of inversions that capture locally adapted alleles when two populations are exchanging migrants or hybridizing. By suppressing recombination between the loci, a new inversion can spread. Neither drift nor coadaptation between the alleles (epistasis) is needed, so this local adaptation mechanism may apply to a broader range of genetic and demographic situations than alternative hypotheses that have been widely discussed. The mechanism can explain many features observed in inversion systems. It will drive an inversion to high frequency if there is no countervailing force, which could explain fixed differences observed between populations and species. An inversion can be stabilized at an intermediate frequency if it also happens to capture one or more deleterious recessive mutations, which could explain polymorphisms that are common in some species. This polymorphism can cycle in frequency with the changing selective advantage of the locally favored alleles. The mechanism can establish underdominant inversions that decrease heterokaryotype fitness by several percent if the cause of fitness loss is structural, while if the cause is genic there is no limit to the strength of underdominance that can result. The mechanism is expected to cause loci responsible for adaptive species-specific differences to map to inversions, as seen in recent QTL studies. We discuss data that support the hypothesis, review other mechanisms for inversion evolution, and suggest possible tests.
Figures



References
-
- Andolfatto, P., F. Depaulis and A. Navarro, 2001. Inversion polymorphisms and nucleotide variability in Drosophila. Genet. Res. 77: 1–8. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

