Adaptive evolution of heparanase in hypoxia-tolerant Spalax: gene cloning and identification of a unique splice variant
- PMID: 16204381
- PMCID: PMC1257723
- DOI: 10.1073/pnas.0507279102
Adaptive evolution of heparanase in hypoxia-tolerant Spalax: gene cloning and identification of a unique splice variant
Abstract
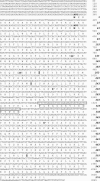
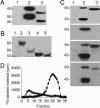
Heparan sulfate (HS) side chains of HS proteoglycans bind to and assemble extracellular matrix proteins and play important roles in cell-cell and cell-extracellular matrix interactions. HS chains bind a multitude of bioactive molecules and thereby function in the control of multiple normal and pathological processes. Enzymatic degradation of HS by heparanase, a mammalian endoglycosidase, affects the integrity and functional state of tissues and is involved in, among other processes, inflammation, angiogenesis, and cancer metastasis. Here, we report the cloning of heparanase from four Israeli species of the blind subterranean mole rat (Spalax ehrenbergi superspecies), 85% homologous to the human enzyme. Unlike its limited expression in human tissues, heparanase is highly expressed in diverse Spalax tissues. Moreover, we have identified a unique splice variant of the Spalax enzyme lacking 16 aa encoded by exon 7. This deletion resulted in a major defect in trafficking and processing of the heparanase protein, leading to a loss of its enzymatic activity. Interspecies variation was noted in the sequence and in the expression of the splice variant of the heparanase gene in blind mole rats living under different ecogeographical stresses, indicating a possible role in adaptation to stress in Spalax evolution.
Figures



References
-
- Kjellen, L. & Lindahl, U. (1991) Annu. Rev. Biochem. 60, 443-475. - PubMed
-
- Bernfield, M., Gotte, M., Park, P. W., Reizes, O., Fitzgerald, M. L., Lincecum, J. & Zako, M. (1999) Annu. Rev. Biochem. 68, 729-777. - PubMed
-
- Iozzo, R. V. (1998) Annu. Rev. Biochem. 67, 609-652. - PubMed
-
- Vlodavsky, I., Bar-Shavit, R., Korner, G. & Fuks, Z. (1993) in Basement Membranes: Cellular and Molecular Aspects, eds. Rohrbach, D. H. & Timpl, R. (Academic, Orlando, FL), pp. 327-343.
-
- Vlodavsky, I., Miao, H.-Q., Medalion, B., Danagher, P. & Ron, D. (1996) Cancer Metastasis Rev. 15, 177-186. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

