Quantification of Campylobacter spp. in chicken rinse samples by using flotation prior to real-time PCR
- PMID: 16204485
- PMCID: PMC1265984
- DOI: 10.1128/AEM.71.10.5759-5764.2005
Quantification of Campylobacter spp. in chicken rinse samples by using flotation prior to real-time PCR
Abstract
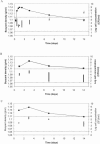
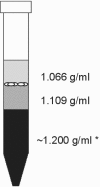
Real-time PCR is fast, sensitive, specific, and can deliver quantitative data; however, two disadvantages are that this technology is sensitive to inhibition by food and that it does not distinguish between DNA originating from viable, viable nonculturable (VNC), and dead cells. For this reason, real-time PCR has been combined with a novel discontinuous buoyant density gradient method, called flotation, in order to allow detection of only viable and VNC cells of thermotolerant campylobacters in chicken rinse samples. Studying the buoyant densities of different Campylobacter spp. showed that densities changed at different time points during growth; however, all varied between 1.065 and 1.109 g/ml. These data were then used to develop a flotation assay. Results showed that after flotation and real-time PCR, cell concentrations as low as 8.6 x 10(2) CFU/ml could be detected without culture enrichment and amounts as low as 2.6 x 10(3) CFU/ml could be quantified. Furthermore, subjecting viable cells and dead cells to flotation showed that viable cells were recovered after flotation treatment but that dead cells and/or their DNA was not detected. Also, when samples containing VNC cells mixed with dead cells were treated with flotation after storage at 4 or 20 degrees C for 21 days, a similar percentage resembling the VNC cell fraction was detected using real-time PCR and 5-cyano-2,3-ditolyl tetrazolium chloride-4',6'-diamidino-2-phenylindole staining (20% +/- 9% and 23% +/- 4%, respectively, at 4 degrees C; 11% +/- 4% and 10% +/- 2%, respectively, at 20 degrees C). This indicated that viable and VNC Campylobacter cells could be positively selected and quantified using the flotation method.
Figures

Similar articles
-
Simultaneous quantification of pathogenic Campylobacter and Salmonella in chicken rinse fluid by a flotation and real-time multiplex PCR procedure.Int J Food Microbiol. 2007 Jun 10;117(1):50-4. doi: 10.1016/j.ijfoodmicro.2007.02.020. Epub 2007 Mar 30. Int J Food Microbiol. 2007. PMID: 17462769
-
Detection of Campylobacter spp. in chicken fecal samples by real-time PCR.J Clin Microbiol. 2004 Nov;42(11):5125-32. doi: 10.1128/JCM.42.11.5125-5132.2004. J Clin Microbiol. 2004. PMID: 15528705 Free PMC article.
-
Risk assessment of false-positive quantitative real-time PCR results in food, due to detection of DNA originating from dead cells.J Microbiol Methods. 2005 Mar;60(3):315-23. doi: 10.1016/j.mimet.2004.10.003. J Microbiol Methods. 2005. PMID: 15649533
-
Detection and identification of Campylobacter spp. using the polymerase chain reaction.Cell Mol Biol (Noisy-le-grand). 1995 Jul;41(5):625-38. Cell Mol Biol (Noisy-le-grand). 1995. PMID: 7580843 Review.
-
Where does Campylobacter come from? A molecular odyssey.Adv Exp Med Biol. 2010;659:47-56. doi: 10.1007/978-1-4419-0981-7_4. Adv Exp Med Biol. 2010. PMID: 20204754 Free PMC article. Review. No abstract available.
Cited by
-
Enrichment Free qPCR for Rapid Identification and Quantification of Campylobacter jejuni, C. coli, C. lari, and C. upsaliensis in Chicken Meat Samples by a New Couple of Primers.Foods. 2021 Sep 30;10(10):2341. doi: 10.3390/foods10102341. Foods. 2021. PMID: 34681388 Free PMC article.
-
Rapid separation and concentration of food-borne pathogens in food samples prior to quantification by viable-cell counting and real-time PCR.Appl Environ Microbiol. 2007 Jan;73(1):92-100. doi: 10.1128/AEM.01772-06. Epub 2006 Oct 20. Appl Environ Microbiol. 2007. PMID: 17056684 Free PMC article.
-
Enumeration of salmonella bacteria in food and feed samples by real-time PCR for quantitative microbial risk assessment.Appl Environ Microbiol. 2008 Mar;74(5):1299-304. doi: 10.1128/AEM.02489-07. Epub 2007 Dec 28. Appl Environ Microbiol. 2008. PMID: 18165357 Free PMC article. Review. No abstract available.
-
An easy-to-perform, culture-free Campylobacter point-of-management assay for processing plant applications.J Appl Microbiol. 2020 Mar;128(3):620-629. doi: 10.1111/jam.14509. Epub 2019 Dec 4. J Appl Microbiol. 2020. PMID: 31705613 Free PMC article.
-
Rapid quantification of viable Campylobacter bacteria on chicken carcasses, using real-time PCR and propidium monoazide treatment, as a tool for quantitative risk assessment.Appl Environ Microbiol. 2010 Aug;76(15):5097-104. doi: 10.1128/AEM.00411-10. Epub 2010 Jun 18. Appl Environ Microbiol. 2010. PMID: 20562292 Free PMC article.
References
-
- Allos, B. M., and M. J. Blaser. 1995. Campylobacter jejuni and the expanding spectrum of related infections. Clin. Infect. Dis. 20:1092-1099. - PubMed
-
- Cappelier, J. M., B. Lazaro, A. Rossero, A. Fernandez-Astorga, and M. Federighi. 1997. Double staining (CTC-DAPI) for detection and enumeration of viable but non-culturable Campylobacter jejuni cells. Vet. Res. 28:547-555. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

