Genetic diversity of Listeria monocytogenes strains from a high-prevalence dairy farm
- PMID: 16204502
- PMCID: PMC1266006
- DOI: 10.1128/AEM.71.10.5893-5899.2005
Genetic diversity of Listeria monocytogenes strains from a high-prevalence dairy farm
Abstract
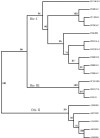
Listeria monocytogenes is a significant food-borne human and veterinary pathogen. Contaminated silage commonly leads to disease in livestock, but the pervasive nature of the bacterium can make it difficult to identify the source of infection. An investigation of bovine listeriosis that occurred on a Pacific Northwest dairy farm ("farm A") revealed that the clinical strain was closely related to fecal strains from asymptomatic cows, and that farm environment was heavily contaminated with a diversity of L. monocytogenes strains. In addition, the farm A clinical strain was closely related to clinical and environmental strains obtained 1 year prior from a second Northwest dairy farm ("farm B"). To investigate the source(s) of contamination on farm A, environmental samples were collected from farm A at two time points. Pulsed-field gel electrophoresis characterization of 538 isolates obtained from that farm identified 57 different AscI pulsovars. Fecal isolates obtained from individual cows were the most genetically diverse, with up to 94% of fecal samples containing more than one pulsovar. The maximum numbers of pulsovars and serotypes isolated from a fecal sample of one cow were 6 and 4, respectively. Serotype 1/2a was isolated most frequently at both time points. Microarray genotyping of bovine listeriosis, fecal, and silage strains from both farms identified four probes that differentiated listeriosis strains from environmental strains; however, no probe was common to both bovine listeriosis strains.
Figures



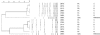
Similar articles
-
Occurrence, Persistence, and Contamination Routes of Listeria monocytogenes Genotypes on Three Finnish Dairy Cattle Farms: a Longitudinal Study.Appl Environ Microbiol. 2018 Jan 31;84(4):e02000-17. doi: 10.1128/AEM.02000-17. Print 2018 Feb 15. Appl Environ Microbiol. 2018. PMID: 29222098 Free PMC article.
-
Diversity of Listeria monocytogenes within a U.S. dairy herd, 2004-2010.Foodborne Pathog Dis. 2015 Oct;12(10):844-50. doi: 10.1089/fpd.2014.1886. Epub 2015 Sep 1. Foodborne Pathog Dis. 2015. PMID: 26325149
-
Molecular diversity of Listeria monocytogenes isolated from Irish dairy farms.Foodborne Pathog Dis. 2011 May;8(5):635-41. doi: 10.1089/fpd.2010.0806. Epub 2011 Jan 19. Foodborne Pathog Dis. 2011. PMID: 21247298
-
The prevalence of Listeria monocytogenes in the environment of dairy farms.Microbiol Immunol. 1996;40(2):121-24. doi: 10.1111/j.1348-0421.1996.tb03326.x. Microbiol Immunol. 1996. PMID: 8867607
-
Listeria monocytogenes lineages: Genomics, evolution, ecology, and phenotypic characteristics.Int J Med Microbiol. 2011 Feb;301(2):79-96. doi: 10.1016/j.ijmm.2010.05.002. Epub 2010 Aug 13. Int J Med Microbiol. 2011. PMID: 20708964 Review.
Cited by
-
Molecular ecology of Listeria monocytogenes: evidence for a reservoir in milking equipment on a dairy farm.Appl Environ Microbiol. 2009 Mar;75(5):1315-23. doi: 10.1128/AEM.01826-08. Epub 2008 Dec 29. Appl Environ Microbiol. 2009. PMID: 19114514 Free PMC article.
-
Rhombencephalitis Caused by Listeria monocytogenes in Humans and Ruminants: A Zoonosis on the Rise?Interdiscip Perspect Infect Dis. 2010;2010:632513. doi: 10.1155/2010/632513. Epub 2010 Feb 28. Interdiscip Perspect Infect Dis. 2010. PMID: 20204066 Free PMC article.
-
Risk factors associated with fecal shedding of Listeria monocytogenes by dairy cows and calves.J Vet Intern Med. 2018 Sep;32(5):1773-1779. doi: 10.1111/jvim.15234. Epub 2018 Sep 17. J Vet Intern Med. 2018. PMID: 30295965 Free PMC article.
-
Transmission Scenarios of Listeria monocytogenes on Small Ruminant On-Farm Dairies.Foods. 2023 Jan 6;12(2):265. doi: 10.3390/foods12020265. Foods. 2023. PMID: 36673359 Free PMC article. Review.
-
Emergence and global spread of Listeria monocytogenes main clinical clonal complex.Sci Adv. 2021 Dec 3;7(49):eabj9805. doi: 10.1126/sciadv.abj9805. Epub 2021 Dec 1. Sci Adv. 2021. PMID: 34851675 Free PMC article.
References
-
- Border, P. M., J. J. Howard, G. S. Plastow, and K. W. Siggens. 1990. Detection of Listeria species and Listeria monocytogenes using polymerase chain reaction. Lett. Appl. Microbiol. 11:158-162. - PubMed
-
- Borucki, M. K., M. J. Krug, W. T. Muraoka, and D. R. Call. 2003. Discrimination among Listeria monocytogenes isolates using a mixed genome DNA microarray. Vet. Microbiol. 92:351-362. - PubMed
-
- Borucki, M. K., J. Reynolds, C. C. Gay, K. L. McElwain, S. H. Kim, D. P. Knowles, and J. Hu. 2004. Dairy farm reservoir of Listeria monocytogenes sporadic and epidemic strains. J. Food Prot. 67:2496-2499. - PubMed
-
- Borucki, M. K., S. H. Kim, D. R. Call, S. C. Smole, and F. Pagotto. 2004. Selective discrimination of Listeria monocytogenes epidemic strains by a mixed-genome DNA microarray compared to discrimination by pulsed-field gel electrophoresis, ribotyping, and multilocus sequence typing. J. Clin. Microbiol. 42:5270-5276. - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

