Specific detection, isolation, and characterization of selected, previously uncultured members of the freshwater bacterioplankton community
- PMID: 16204504
- PMCID: PMC1265938
- DOI: 10.1128/AEM.71.10.5908-5919.2005
Specific detection, isolation, and characterization of selected, previously uncultured members of the freshwater bacterioplankton community
Abstract
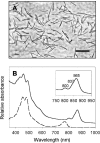
High-throughput cultivation was combined with rapid and group-specific phylogenetic fingerprinting in order to recover representatives of three freshwater bacterioplankton communities. A total of 570 bacterial cultures were obtained by employing the most probable number and MicroDrop techniques. The majority of the cultured bacteria were closely related to previously uncultured bacteria and grouped with the alpha-Proteobacteria, beta-Proteobacteria, Actinobacteria, Firmicutes, or Flavobacteria-Cytophaga lineage. Correspondingly, the natural bacterioplankton community was analyzed by high-resolution phylogenetic fingerprinting of these five bacterial lineages. 16S rRNA gene fragments were generated for each lineage and subsequently separated by denaturing gradient gel electrophoresis. By the combination of five group-specific PCR protocols, the total number of 16S rRNA gene fingerprints generated from the natural communities was increased sixfold compared to conventional (eubacterial) fingerprinting. Four of the environmental alpha-Proteobacteria 16S rRNA gene sequences obtained from the natural community were found to be identical to those of bacterial isolates. One of these phylotypes was detected in 14 different cultures and hence represented the most frequently cultured bacterium. Three of these 14 strains were characterized in detail. Their complete 16S rRNA gene sequences showed only 93% similarity to that of Sandaracinobacter sibiricus, the closest relative described so far. The novel phylotype of bacterium is a strict aerobe capable of using numerous organic carbon substrates and contains bacteriochlorophyll a bound to two different photosynthetic light-harvesting complexes. Dot blot hybridization revealed that the strains occur in lakes of different trophic status and constitute up to 2% of the microbial community.
Figures



References
-
- Acinas, S. G., V. Klepac-Ceraj, D. E. Hunt, C. Pharino, I. Ceraj, D. L. Distel, and M. F. Polz. 2004. Fine-scale phylogenetic architecture of a complex bacterial community. Nature 430:551-554. - PubMed
-
- Bartscht, K., H. Cypionka, and J. Overmann. 1999. Evaluation of cell activity and of methods for the cultivation of bacteria from a natural lake community. FEMS Microbiol. Ecol. 28:249-259.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

